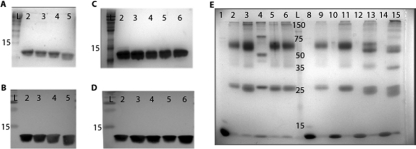
FIG. 2.
Purified AmrZ forms oligomers in solution, and oligomerization does not require the first 26 residues. (A and B) Purification and verification of the wild type and the extended amino truncation proteins Δ2AmrZ, Δ5AmrZ, and Δ11AmrZ by SDS-PAGE and visualization by GelCode staining (A) and Western blotting (B). Lane 2, wild-type AmrZ; lane 3, Δ2AmrZ; lane 4, Δ5AmrZ; lane 5, Δ11AmrZ. (C and D) Purification and verification in parallel of the wild type and the R14A AmrZ (lane 3), K18A AmrZ (lane 4), V20A AmrZ (lane 5), and R22A AmrZ (lane 6) β-sheet mutants by SDS-PAGE and visualization by GelCode staining (C) and Western blotting (D). (E) Samples were cross-linked by incubation with glutaraldehyde to form a stable complex that would withstand separation by SDS-PAGE. Lanes 1 to 6 are the β-sheet mutants. AmrZ in lane 1 was not incubated with glutaraldehyde to indicate the size of a wild-type AmrZ monomer. Lanes 2 to 6 are cross-linked and contain wild-type AmrZ (lane 2), R14A AmrZ (lane 3), K18A AmrZ (lane 4), V20A AmrZ (lane 5), and R22A AmrZ (lane 6). Lanes 8 to 15 represent the truncation proteins with alternating untreated monomeric and glutaraldehyde-treated samples. Lanes 8 and 9, wild-type AmrZ; lanes 10 and 11, Δ2AmrZ; lanes 12 and 13, Δ5AmrZ; lanes 14 and 15, Δ11AmrZ. All lanes contain 40 μmol of protein and were separated by SDS-PAGE on a 12% polyacrylamide gel, and complexes were visualized by GelCode staining.