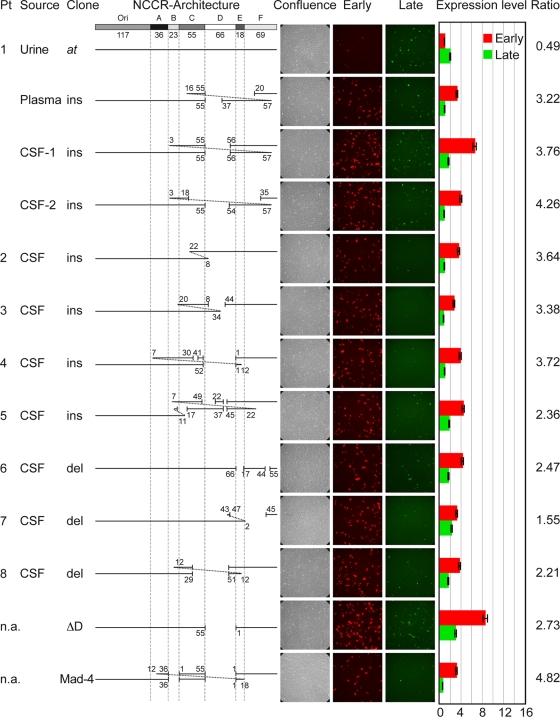
FIG. 1.
JCV NCCR architecture and reporter gene expression in PDA cells. The at-NCCR of JCV is shown with the following arbitrarily denoted blocks (the number of base pairs is in parentheses): Ori(117)-A(36)-B(23)-C(55)-D(66)-E(18)-F(69). Also shown is the architecture of the rr-NCCRs. Pt, patient; source, origin of sample; ins, insertions corresponding to duplications of the numbered base pairs; del, deletions denoted by gaps and nucleotide number; confluence, samples showing phase contrast; early, red fluorescence; late, green fluorescence. Cells were transfected with the indicated bidirectional reporter constructs, and expression was quantified at 2 dpt. Expression level indicates the fold induction of green and red fluorescence-positive cells normalized to GFP- and RFP-positive cells bearing pHRG/urine at the at-NCCR; ratio, the number of RFP-positive cells divided by the number of GFP-positive cells; n.a., not applicable, since the NCCR has been generated by in vitro mutagenesis or represents the NCCR of reference strain Mad-4. Means from three independent experiments are shown. Error bars represent the means ± standard deviations.