FIG. 1.
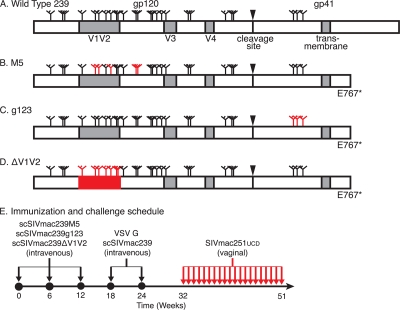
Animals were immunized with Env-modified single-cycle SIV and challenged by repeated, low-dose vaginal inoculation. A schematic representation of the wild-type SIVmac239 Env appears at the top (A). Positions of potential sites of N-linked glycosylation (N-X-S/T motifs) are indicated by tree-like symbols. Features removed from each Env-modified strain are indicated (red). The M5 Env has asparagine-to-glutamine substitutions that eliminate the 5th, 6th, 8th, 12th, and 13th potential N-linked glycosylation sites in gp120 (B). The g123 Env has all three potential N-linked glycosylation sites in gp41 similarly eliminated (C). Variable loops 1 and 2 of the ΔV1V2 envelope are deleted (D). All of the modified envelopes have a glutamate-to-stop codon change at position 767 (E767*). Eight macaques received an intravenous injection consisting of a mixture of 5 μg p27 equivalents of scSIVmac239M5, scSIVmac239g123, and scSIVmac239ΔV1V2 on weeks 0, 6, and 12 (E). All eight animals also received 15 μg p27 equivalents of VSV G trans-complemented scSIVmac239 intravenously on weeks 18 and 24. Beginning on week 32, the animals were challenged vaginally with 1,000 TCID50 (1 ng p27) of SIVmac251UCD twice per day on the same day each week for 20 weeks or until viral RNA was detected in plasma on two consecutive weeks.