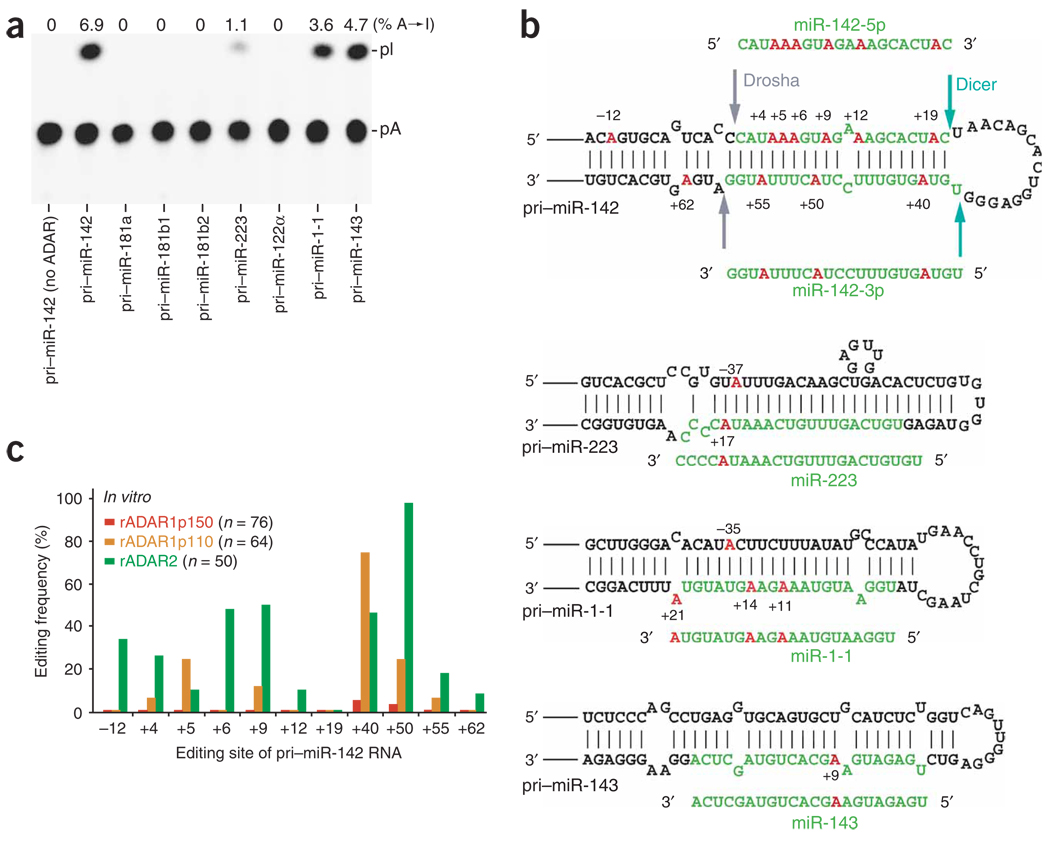
Figure 1.
A→I RNA editing of pri-miR RNAs by ADAR. (a) TLC analysis of pri-miRNAs edited in vitro by a mixture of recombinant ADAR1p110 and ADAR2 proteins (rADAR1p110 and rADAR2, 20 ng each). (b) A→I editing sites of four pri-miRNAs. Red, edited adenosine residues (numbered by position with the 5′ end of the mature miRNA sequence counted as +1); green, the region to be processed into the mature miRNA; arrows in pri–miR-142 hairpin structure, cleavage sites for Drosha and Dicer. (c) A quantitative summary of the editing patterns revealed by sequencing of RT-PCR cDNA clones corresponding to pri–miR-142 RNAs edited in vitro by rADAR1p150, rADAR1p110 or rADAR2. Editing frequency is represented as a percentage (number of independent cDNA clones representing the edited pri–miR-142 sequence at that site divided by the total number of cDNA clones examined).