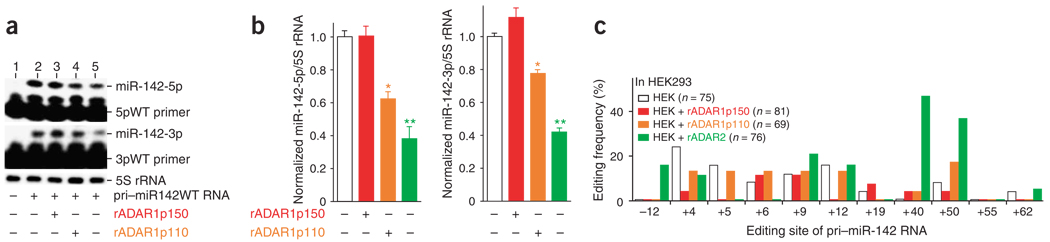
Figure 4.
Suppression of pri–miR-142 processing in HEK293 cells transfected with ADAR expression plasmids. (a) Inhibitory effects of ADAR1p110 and ADAR2 on processing of pri–miR-142 in HEK293 cells. Primer-extension assays monitoring for expression levels of miR-142-5p (upper gel) and miR-142-3p (lower gel) as in Figure 2d are shown. (b) Histograms representing data like that in a from three independent experiments. The relative miR-142-5p and miR-142-3p levels in HEK293 cells cotransfected with the ADAR1p150, ADAR1p110 or ADAR2 expression plasmid were compared statistically by individual unpaired Student’s t-tests to those in HEK293 cells transfected only with pri-miR142WT plasmid. Significant differences are indicated as follows: one asterisk, P < 0.01; two asterisks, P < 0.001. Error bars, s.e.m. (c) A quantitative summary (as in Figure 1c) of the editing pattern revealed by sequencing of cDNA clones corresponding to pri–miR-142 RNAs edited in transfected HEK293 cells. White bars represent the results of experiments conducted with HEK 293 cells transfected solely with wild-type pri–miR-142 RNA expression plasmid.