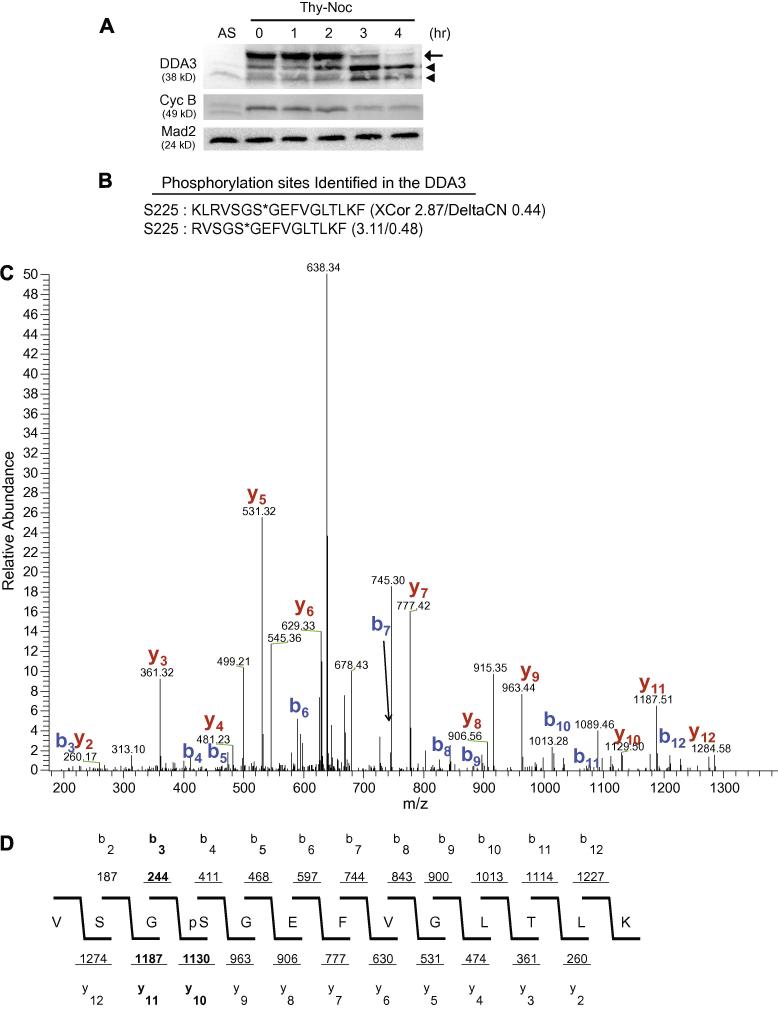
Figure 1. DDA3 is phosphorylated at Ser225 in mitosis.
(A) HeLa cells were synchronized by a thymidine-nocodazole (Thy-Noc) block, released into fresh media and harvested at indicated time. The arrow points to phosphorylated mitotic DDA3 and arrowheads point to two forms of interphase DDA3, as characterized previously [6]. AS, unsynchronized cells. (B-D) DDA3-S-GFP cells were synchronized at prometaphase by a thymidine-nocodazole arrest and the DDA3 protein was tandem-affinity-purified from the mitotic DDA3-S-GFP cells and analyzed by mass spectrometry. Peptides derived from phosphorylated DDA3 are listed in (B) with XCorr and DeltaCN scores indicated (*, phosphorylation site). Shown in (C) was the actual spectrum for the second peptide listed in (B) with signals for the b-ion and y-ion series indicated. The spectrum data was summarized in (D). The validity of the phospho-peptide VSGS*GEFVGLTLK (C & D) was confirmed by the software DeBunker [14]. Manual annotation of the tandem mass spectrum also confirmed the phosphorylation at S225. Of the four site-determining ions for Ser225, b3, y10 and y11 [highlighted in (D)] were observed in the tandem mass spectra, providing great confidence on the validity of this phosphorylation site. The fourth site-determining ion b2 was not within the scanning range of the instrument.