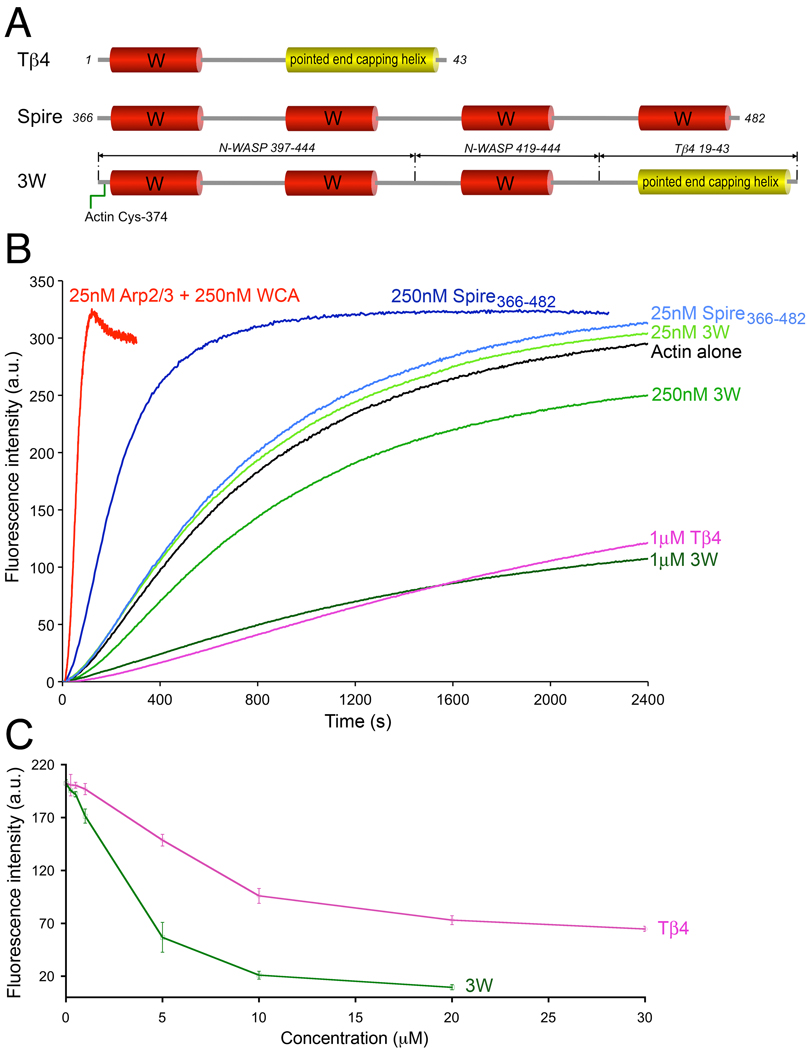
FIG. 5.
Different effects of Spire, 3W, and Tβ4 on actin polymerization. (A) Schematic diagram of Tβ4, the four W domain region of Drosophila Spire, and construct 3W. Note that construct 3W consists of three W domains (occurring naturally in mouse N-WASP) separated by short linkers (as in Spire) and the pointed end capping helix of Tβ4. This construct also contains a Cys residue at the N-terminus that was crosslinked to actin Cys-374 for crystallization, but the crosslink was reduced with DTT to measure the nucleation activity. (B) Time course of polymerization of 2 µM Mg-ATP-actin (6% pyrene-labeled) alone (black) or in the presence of different concentrations of Spire366–482 (different shades of blue), construct 3W (different shades of green), Tβ4 (pink), and 25 nM Arp2/3 complex with 250 nM mouse N-WASP WCA (red). Each experiment was repeated at least three times. The polymerization rates are: Actin (0.8±0.1 nM/sec), Spire (1.0±0.2 nM/sec at 25nM and 4.8±0.2 nM/sec at 250 nM), Arp2/3 complex (31.5±1 nM/sec). (C) Steady-state concentration-dependence of actin monomer sequestration by Tβ4 and 3W. (One Column Figure)