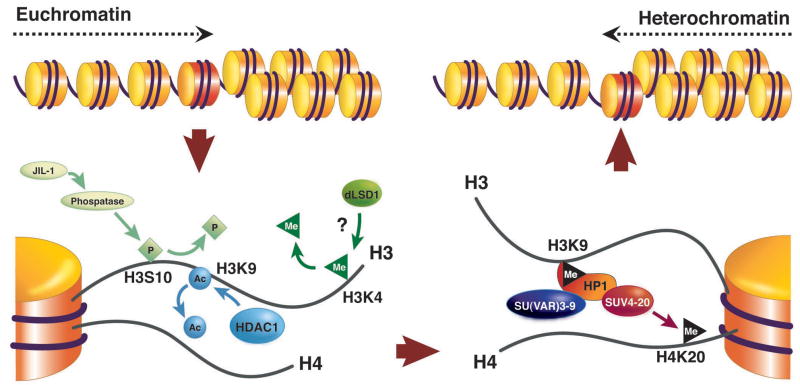
Figure 1.
Changes in histone modification implicated in the switch from a euchromatic to a heterochromatic state. Active genes are frequently marked by H3K4me (but see Berger, this issue); this mark must presumably be removed by LSD1 (not yet characterized in Drosophila). H3K9 is normally acetylated in euchromatin, and this mark must be removed by a histone deacetylase, typically HDAC1. Phosphorylation of H3S10 can interfere with methylation of H3K9; dephosphorylation may involved a phosphatase targeted through the carboxyl terminus of the JIL1 kinase10. These transitions set the stage for acquisition of the modifications associated with silencing, including methylation of H3K9 by SU(VAR)3-9 or another HMT, binding of HP1, and subsequent methylation of H4K20 by SUV4-20, an enzyme recruited by HP1. Other silencing marks, such as methylation of H3K27 by E(Z) (not shown), appear to be relevant in some domains, although this mark is more prominently used by the Polycomb system. Supporting data comes from genetic identification of modifiers of PEV, biochemical characterization of the activities of such modifiers, and tests of protein-protein interactions 9. (Adapted from10)