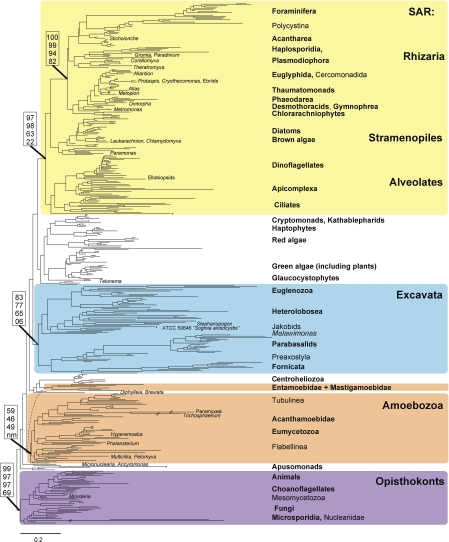
FIGURE 1.
Most likely eukaryotic tree of life reconstructed using all 451 taxa and all 16 genes (SSU-rDNA plus 15 protein genes). Major nodes in this topology are robust to analyses of subsets of taxa and genes, which include varying levels of missing data (Table 1). Clades in bold are monophyletic in analyses with 2 or more members except in all:15 in which taxa represented by a single gene were sometimes misplaced. Numbers in boxes represent support at key nodes in analyses with increasing amounts of missing data (10:16, 6:16, 4:16, and all:16 analyses; see Table 1 for more details). Given uncertainties around the root of the eukaryotic tree of life (see text), we have chosen to draw the tree rooted with the well-supported clade Opisthokonta. Dashed line indicates alternate branching pattern seen for Amoebozoa in other analyses. Long branches, indicated by //, have been reduced by half. The 6 lineages labeled by * represent taxa that are misplaced, probably due to LBA, listed from top to bottom with expected clade in parentheses. These are Protoopalina japonica (Stramenopiles), Aggregata octopiana (Apicomplexa), Mikrocytos mackini (Haplosporidia), Centropyxis laevigata (Tubulinea), Marteilioides chungmuensis (unplaced), and Cochliopodium spiniferum (Amoebozoa).