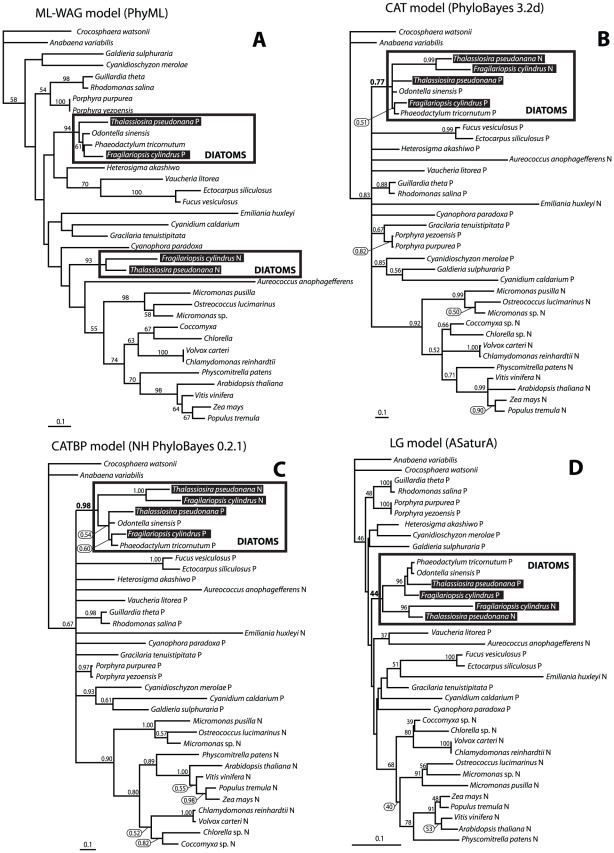
Figure 3. Phylogenetic trees as inferred from amino acid sequences of Psb28 (112 residues).
Trees were constructed using conventional maximum likelihood (A) and methods which can deal with a compositional bias of the sequences and phylogenetic artefacts such as LBA as demonstrated here. PhyloBayes 3.2d (CAT model) (B) and NHPhyloBayes (CAT-BP model for different evolutionary rates among taxa) (C) and AsaturA (designed to deal with amino acid saturation) (D) show the expected origin of nuclear encoded Psb28 from T. pseudonana. It appears that psb28 was transferred to the nucleus through endosymbiotic transfer from the diatom plastid to the diatom nucleus. Numbers above branches show Bayesian posterior probabilities (PhyloBayes 3.2d: 100000 steps, burnin = 20000, trees sampled every tenth step; HN PhyloBayes 0.2.1: 10000 steps, burnin = 2000, trees sampled each step) and bootstraps (AsaturA; 1000 replicates).