Abstract
Heat shock protein 70 (Hsp70) is a highly conserved molecular chaperone that plays multiple roles in protein homeostasis. In these various tasks, the activity of Hsp70 is shaped by interactions with co-chaperones, such as Hsp40. The Hsp40 family of co-chaperones binds to Hsp70 through a conserved J-domain, and these factors stimulate ATPase and protein-folding activity. Using chemical screens, we identified a compound, 115-7c, which acts as an artificial co-chaperone for Hsp70. Specifically, the activities of 115-7c mirrored those of a Hsp40; the compound stimulated the ATPase and protein-folding activities of a prokaryotic Hsp70 (DnaK) and partially compensated for a Hsp40 loss-of-function mutation in yeast. Consistent with these observations, NMR and mutagenesis studies indicate that the binding site for 115-7c is adjacent to a region on DnaK that is required for J-domain-mediated stimulation. Interestingly, we found that 115-7c and the Hsp40 do not compete for binding but act in concert. Using this information, we introduced additional steric bulk to 115-7c and converted it into an inhibitor. Thus, these chemical probes either promote or inhibit chaperone functions by regulating Hsp70–Hsp40 complex assembly at a native protein–protein interface. This unexpected mechanism may provide new avenues for exploring how chaperones and co-chaperones cooperate to shape protein homeostasis.
Heat shock protein 70 (Hsp70) is a member of a ubiquitously expressed family of molecular chaperones that are involved in protein homeostasis. In its role as a mediator of protein fate, this chaperone has been linked to multiple tasks, including roles in de novo protein folding, subcellular trafficking, protein disaggregation, proteasome-mediated degradation, and autophagy (1–6). In addition, Hsp70 has been linked to numerous diseases, especially cancer and disorders of protein folding (7). Thus, there is interest in better understanding the biology of Hsp70 in order to test its potential as a therapeutic target (8).
To accomplish its various chaperone functions, Hsp70 physically interacts with the exposed hydrophobic residues of polypeptides via its C-terminal substrate-binding domain (SBD). Hydrolysis of ATP in the adjacent, N-terminal nucleotide-binding domain (NBD) propagates an allosteric change to the SBD, resulting in an approximately 10-fold enhancement in substrate affinity (9–12). These findings suggest an important role for the nucleotide state in controlling the interactions of Hsp70 with misfolded substrates. Consistent with the proposed importance of nucleotide turnover, a family of essential co-chaperones, the Hsp40s, is known to tightly regulate the ATPase rate of Hsp70. These co-chaperones are defined by the presence of a conserved, 60 amino acid J-domain. Interaction of the J-domain with the NBD of a Hsp70 stimulates its ATP hydrolysis and favors tight association with bound substrates. For example, the J-domain containing co-chaperone, DnaJ, stimulates the nucleotide hydrolysis rate of the well-characterized prokaryotic Hsp70, DnaK, by at least 5-fold and favors DnaK-substrate interactions (13–16). The residues in the NBD that are important for this co-chaperone partnership are ~15–20 Å removed from the nucleotide-binding cleft, suggesting that the J-domain controls ATPase activity through allostery (17–20). Regulation by J-domain-containing Hsp40s appears to be critical, because loss-of-function mutations compromise viability in vivo (21, 22). Moreover, overexpression of Hsp40s is sometimes sufficient to impact disease in certain models, further suggesting an important physiological role for the Hsp70–Hsp40 complex (23, 24). Finally, most organisms express multiple J-domain proteins (e.g., more than 40 examples are known in humans and approximately 22 in Saccharomyces cerevisiae), which is an observation that has led to the hypothesis that some co-chaperones play specific cellular roles, such as during a stress response (22, 25–27). Despite these important advances and interesting observations, one of the major questions in chaperone biology is how Hsp70 and its Hsp40 co-chaperones cooperate to shape specific cellular responses.
To understand chaperone regulation in greater detail, we have designed chemical screens against the multiprotein complex (i.e., Hsp70 and a representative J-domain co-chaperone) to uncover probes that might be used to control Hsp70 within the context of the intact chaperone machine (7, 28–31). During these studies, we found that certain dihydropyrimidines impact the ATPase activity of Hsp70, but only in the presence of J-domain proteins (32). For example, the dihydropyrimidine MAL3-101 inhibits ATPase activity, but only when added together with the J-domain of T-antigen (32). These findings suggest an interesting but mysterious interplay between this class of compounds and the Hsp40 co-chaperones. It appears to be worthwhile to understand this mechanism, because dihydropyrimidines have interesting biological activity in cell-based and animal models of tauopathy (33) and in anticancer studies (31, 34, 35). Here, we present the surprising finding that a model dihydropyrimidine, 115-7c, binds to a region adjacent to the surface required for J-domain stimulation and that it utilizes a parallel allosteric conduit to synergistically regulate Hsp70-dependent ATP turnover. These observations provide insight into the regulation of the chaperone complex and illustrate how chemical probes with interesting and unexpected mechanisms can be uncovered in screens against multiprotein targets.
RESULTS AND DISCUSSION
Identification of a Compound That Stimulates the in Vitro Functions of DnaK
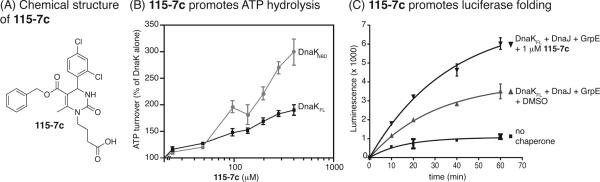
Using ATPase and protein folding activities as end points, we previously identified dihydropyrimidines that either stimulate or inhibit chaperone functions in vitro (29, 30). From these studies, structure–activity analysis led to the synthesis of 115-7c (Figure 1, panel A), which is a compound that stimulates the ATPase activity of full-length Escherichia coli Hsp70, DnaK (DnaKFL), by ~50% at 100 μM (Figure 1, panel B). Because the binding of peptide substrates to the SBD of DnaK is known to stimulate ATP hydrolysis (29), our first hypothesis was that 115-7c might also bind in this region. To test this model, we determined the activity of 115-7c against a truncated mutant composed of only the NBD (DnaKNBD). These experiments revealed that 115-7c retained full activity against DnaKNBD (hydrolysis rate increased ~2-fold at 100 μM), suggesting that the SBD is dispensable.
Figure 1.
Compound 115-7c stimulates the ATPase and folding activities of Hsp70-family chaperones. A) Chemical structure of the dihydropyrimidine, 115-7c. B) 115-7c promotes the ATPase activity of E. coli DnaK (DnaKFL) and a truncated DnaK containing only the nucleotide-binding domain (DnaKNBD). Results are the average of triplicates, and the error bars represent the standard deviations. The concentration of proteins: DnaK (0.6 μM), DnaJ (1.0 μM), GrpE (0.9 μM). C) 115-7c promotes the luciferase-folding activity of the DnaKFL–DnaJ–GrpE complex. Results are the average of triplicates and the error bars represent the standard deviations. The concentration of proteins: DnaK (0.2 μM), DnaJ (0.05 μM), GrpE (0.02 μM).
One of the major functions of Hsp70 is to assist in protein folding. Accordingly, the ATP-dependent, chaperone-mediated refolding of chemically denatured firefly luciferase is a common assay for studying this system in vitro (30, 36, 37). To explore the effects of 115-7c in this model, we denatured firefly luciferase with guanidinium hydrochloride and then diluted this substrate in the presence of the prokaryotic Hsp70 system, composed of DnaK, DnaJ, and the nucleotide exchange factor, GrpE. In this platform, 115-7c stimulated refolding activity by ~2-fold at 1 μM (Figure 1, panel C). Together, these results indicate that the dihydropyrimidine, 115-7c, stimulates two major functions of a Hsp70 chaperone system in vitro, ATPase rate and protein folding.
Compound 115-7c Is an Artificial Co-chaperone in Yeast
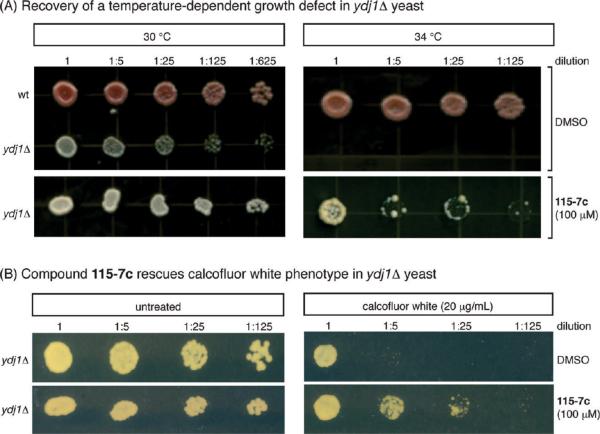
Stimulation of Hsp70s ATPase and folding activities is a function typically associated with J-domain co-chaperones; thus, we hypothesized that 115-7c might act as an artificial co-chaperone in cells. To explore this idea, we turned to a S. cerevisiae model in which a role for J-domains has been clearly documented (22). Ydj1 is one of the DnaJ orthologs in budding yeast and the growth of a ydj1Δ mutant is severely compromised at elevated temperature (21, 22, 38). Based on the in vitro studies, we hypothesized that 115-7c might partially compensate for the loss of YDJ1. To investigate this possibility, we treated ydj1Δ cells with 115-7c by incorporating this compound into the solid growth media. As expected, the mock treated ydj1Δ cells grew slowly at 30 °C and failed to thrive at 34 °C (Figure 2, panel A). Application of 115-7c (100 μM), however, greatly enhanced growth at the elevated temperature (Figure 2, panel A). As an independent test, we also treated ydj1Δ cells with calcofluor white (Figure 2, panel B). Calcofluor white is known to impair cell wall biogenesis and it preferentially decreases the viability of ydj1Δ mutants (39). Treatment with 115-7c (100 μM) partially suppressed this phenotype (Figure 2, panel B), mirroring the results of the thermal stress experiments. We found these results particularly striking because we are unaware of similar examples in which a gene deletion can be recovered by a small, synthetic mimetic. However, it is worth noting that 115-7c only partially compensated for the loss of Ydj1p and relatively high quantities of the small molecule were required.
Figure 2.
Compound 115-7c partially circumvents loss-of-function mutations in the J-domain containing co-chaperone, Ydj1, in yeast. A) Addition of 115-7c (100 μM), but not the solvent control, partially restores growth at elevated temperatures in a ydj1Δ strain. B) Deletion of Ydj1 sensitizes cells to treatment with calcofluor white, and application of 115-7c (100 μM) partially suppresses this phenotype.
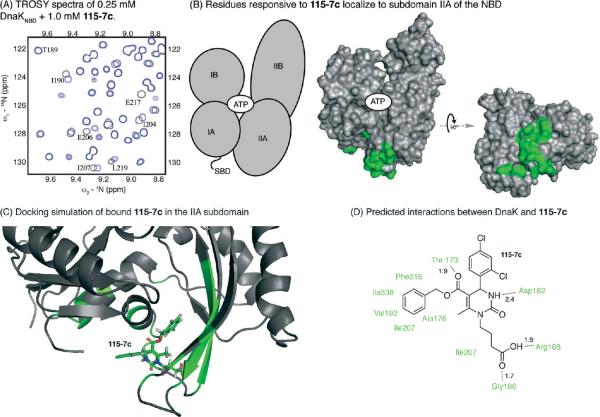
115-7c Binds to the IIA Subdomain of DnaK. To better understand the molecular mechanisms by which 115-7c stimulates ATPase activity and suppresses ydj1Δ phenotypes, we performed nuclear magnetic resonance (NMR) experiments on DnaKNBD. In these experiments, 1H–15N labeled DnaKNBD (250 μM) was titrated with 115-7c and a two-dimensional 15N–1H HSQC-TROSY experiment was performed (Figure 3, panel A) (40, 41). A small, discrete set of chemical shift changes were observed upon 115-7c binding, and these residues largely clustered in the IIA subdomain (Figure 3, panel B and Supplementary Figure 1A). A few residues in the adjacent IA subdomain were also responsive to the compound. Essentially the same pattern of chemical shift changes was observed when 115-7c was titrated into either ADP- or ATP-bound DnaKNBD (Supplementary Figure 1A,B). Moreover, similar though less robust patterns were observed with 15N–1H-labeled DnaKFL (Supplementary Figure 1C), consistent with the results of the ATPase assays (see Figure 1, panel B). Guided by the NMR results, we used Langevin dynamics simulations and induced fit docking in GLIDE to build a model of the 115-7c-bound complex. The best-scored configuration inserted the compound into a hydrophobic cleft at the bottom of the IIA subdomain (Figure 3, panel C). This model included a number of favorable interactions, including the juxtaposition of the carboxylic acid of 115-7c with R188 and the amide nitrogen of G186 and the placement of 115-7c's phenyl group into a deep hydrophobic pocket composed of F216, I207, V192, I338, and A176 (Figure 3, panel D). This orientation is consistent with the NMR chemical shift perturbations and provides a model for how 115-7c might bind the IIA subdomain.
Figure 3.
Compound 115-7c interacts with the subdomain IIA site on DnaK's NBD. A) A subset of the 2D spectra of DnaKNBD (+ 5 mM ATP) is shown in the presence (blue) and absence (gray) of 4 equiv of 115-7c. B) Mapping the chemical shift above 0.02 ppm revealed a localized region at the IIA subdomain. The subdomains of the NBD are shown in the schematic, and the responsive residues are shown in green. C) Model for binding of 115-7c to the IIA subdomain. See the Methods section for details. D) On the basis of the docked configuration, the key interactions between DnaK and 115-7c are predicted. Approximate distances are given in angstroms.
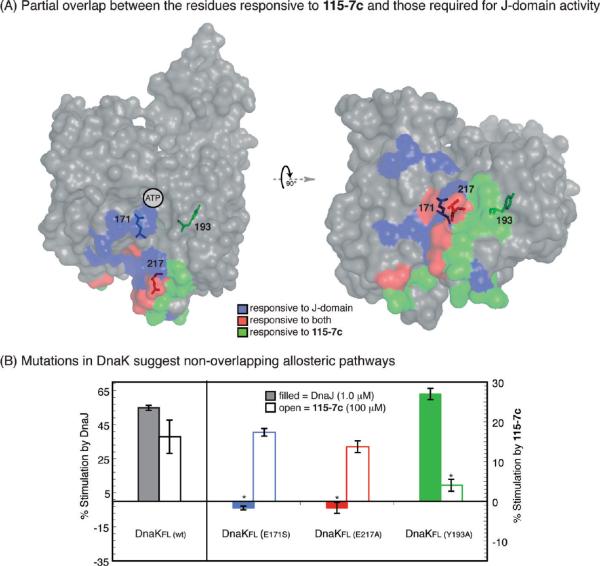
115-7c and DnaJ Appear To Access Parallel Allosteric Mechanisms. Intriguingly, the IA and IIA regions of the NBD have also been implicated in the activity of J-domains, and multiple point mutants in these regions have been found to block stimulation by these co-chaperones (17, 18, 42, 43). To probe whether there might be overlap between these J-domain-responsive residues and those that we found in the NMR experiment with 115-7c, we first highlighted the key amino acids on the structure of DnaKNBD (Figure 4, panel A). This analysis suggested that the artificial and natural co-chaperones occupy only partially overlapping sites. Thus, we hypothesized that mutations in select residues might block allosteric activation by either the J-domain co-chaperone or 115-7c. To test this model, we made point mutations in three residues (E171S, E217A, and Y193A) of DnaKFL and measured the responsiveness of the corresponding mutant proteins. The E171S mutation was chosen because it was previously reported to be required for stimulation by J-domains (17), and consistent with that idea, we confirmed that the ATPase activity of DnaKFl (E171S) was nonresponsive to DnaJ addition (Figure 4, panel B). In contrast, 115-7c retained full activity against this mutant. Thus, these results showed that activation of ATP hydrolysis by the chemical probe and the Hsp40 were separable, suggesting incomplete overlap between the allosteric pathways of these stimulatory factors. In support of this model, we found that DnaKFL (Y193A) was responsive to DnaJ but not 115-7c (Figure 4, panel B). Finally, we explored the influence of a residue that was predicted to be important for both J-domains and 115-7c. This mutant, DnaKFL (E217A), was stimulated by the small molecule but not by DnaJ, further suggesting that the two partners access distinct pathways (Figure 4, panel B). Importantly, although some of these mutations altered the intrinsic hydrolysis rates (17), they did not impact global structure, as measured by circular dichroism (CD; Supplementary Figure 2). Together, the results of these mutagenesis studies suggest that there are at least two allosteric pathways in DnaKNBD and that the artificial and native co-chaperones access parallel conduits. As discussed in more detail below, this activity is likely a byproduct of the screening approach that produced 115-7c; successful compounds were those that stimulated the DnaK–DnaJ complex (29).
Figure 4.
Site-directed mutagenesis suggests at least two allosteric pathways that stimulate the ATPase activity of DnaK. A) Overlay of the residues identified by NMR as responsive to 115-7c (green), those known to be important for J-domain activity (purple), and those amino acids that fit in both categories (red). The J-domain responsive residues were assembled from published studies in which point mutants in Hsp70s were found to block J-domain-mediated stimulation (17, 18, 42, 43). Two views are shown to highlight the fact that some of the responsive residues, such as E171 and Y193, are found in the protein interior, whereas others, such as E217, are surface residues. The overlay was generated in PyMol. B) Point mutants were generated in DnaKFL at either E171S, E217A, or Y193A. The Y193A mutation blocked 115-7c-mediated stimulation but did not completely prevent J-domain activity (p < 0.0001), whereas the other mutations blocked stimulation by DnaJ. Results are the average of (at least) triplicates and error bars represent the standard deviations.
Compound 115-7c Binds Better to the DnaK-DnaJ Complex than to DnaK Alone
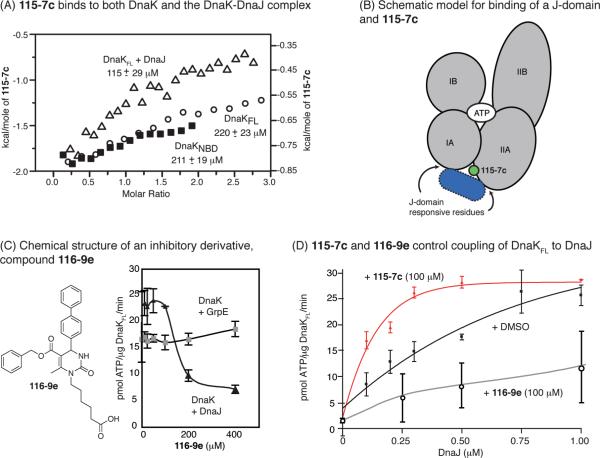
Because the binding sites of the two co-chaperones were only partially overlapping, we wanted to understand whether 115-7c and DnaJ could compete for binding to DnaK. Toward this goal, we first measured 115-7c binding by isothermal calorimetry (ITC) and found that it binds weakly to both DnaKFL (KD ≈ 220 ± 23 μM) and DnaKNBD (KD ≈ 211 ± 19 μM; Figure 5, panel A and Supplementary Figure 3). Next, we attempted to block interactions with 115-7c using saturating levels of DnaJ. Surprisingly, we found that 115-7c bound ca. 2-fold better to the DnaK–DnaJ complex than the isolated chaperone (KD ≈ 113 ± 29 μM; Figure 5, panel A). Because 115-7c had a weak (>1 mM) affinity DnaJ (Supplementary Figure 3B), these results suggest that 115-7c has a more favorable binding site on the composite DnaK–DnaJ complex than on DnaK alone.
Figure 5.
Compound 115-7c and DnaJ bind together to DnaK. A) Isothermal calorimetry reveals that 115-7c binds approximately 2-fold tighter to the DnaK-DnaJ complex than to DnaK alone. Results are representative of duplicate experiments. B) On the basis of the model of bound 115-7c and the ITC results, both DnaJ and 115-7c might bind to DnaK simultaneously. C) Chemical structure of compound 116-9e, which has a diphenyl substitution in place of the solvent-exposed dichlorobenzyl group. This replacement is predicted to sterically interfere with J-domain interactions. 116-9e inhibits nucleotide hydrolysis by the DnaK-DnaJ complex but does not impact GrpE-mediated ATPase activity. Results are the average of triplicates, and the error bars represent standard deviations. D) In the ATPase assay, 115-7c promotes DnaJ-mediated stimulation, whereas 116-9e inhibits activation. Results are the average of triplicate experiments, and the error bars represent standard deviations.
Converting 115-7c into an Inhibitor by Preventing Synergy with the J-domain
On the basis of these results, we envisioned a simple ternary model in which both 115-7c and DnaJ bind DnaK in the cleft between the IIA and IA subdomains (Figure 5, panel B). Because the residues important for these functions are in relatively close proximity, we next explored whether adding bulk to the small molecule could create steric clashes. In these studies, we focused on the dicholorobenzyl moiety of 115-7c, because this region was predicted to be relatively solvent exposed and pointed toward the J-domain responsive residues in the docked model (see Figure 3, panel C). This analysis led to the selection of 116-9e (29), which is similar to 115-7c except that the dichlorobenzyl functionality has been replaced with a bulkier diphenyl group. In the ATPase assay, 116-9e had little effect on the DnaK-GrpE combination, but this agent inhibited the DnaK–DnaJ complex by ~80%, suggesting that it selectively interfered with J-domain-mediated activation of DnaK's ATPase activity (Figure 5, panel C). To further explore this relationship, we titrated DnaJ into DnaKNBD in the presence of either 115-7c or 116-9e and measured ATPase activity. In these experiments, we found that 115-7c reduced the amount of DnaJ required to stimulate DnaKNBD by 1.5-fold (Figure 5, panel D). Conversely, 116-9e strongly suppressed J-domain-mediated stimulation of ATPase activity (Figure 5, panel D). Together, these results suggest that 115-7c and 116-9e tune chaperone activity in the context of the DnaK–DnaJ complex. In further support of this mechanism, 116-9e suppressed DnaK-mediated luciferase folding (Supplementary Figure 4C), which is a process that requires a Hsp40 (30). Together, these results suggest a mechanism in which 115-7c uses an allosteric pathway at a protein–protein interface to stimulate ATPase activity, whereas 116-9e might block DnaJ–DnaK interactions.
Compound 115-7c Phenocopies Hsp70 Overexpression in a Yeast Model of Polyglutamine Disease
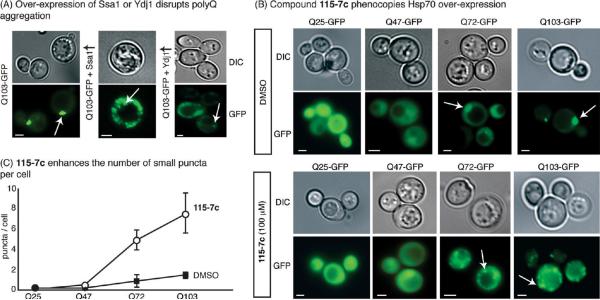
On the basis of the findings that the artificial and natural co-chaperone can sometimes work together, we expected that treatment with 115-7c might stimulate Hsp70 in cells, even those that express a full complement of native co-chaperones. To test this hypothesis, we employed a yeast-based, polyglutamine (polyQ) aggregation model. In this system, expression of an expanded polyQ domain fused to GFP (e.g., Q72- and Q103-GFP) results in fluorescent puncta, while the shorter Q25-GFP and Q47-GFP proteins are less prone to aggregation and therefore remain more diffusely distributed (Figure 6, panel B) (44). We chose this system because overexpression of human Hsp70 has been shown to reduce the tendency of Q103-GFP to form single puncta (44). Thus, we tested whether the stimulatory molecule 115-7c might also produce a similar result. In our hands, overexpression of either SSA1 (a yeast Hsp70 ortholog) or YDJ1 (a yeast Hsp40) altered the fluorescence patterns in Q72- or Q103-expressing cells, resulting in multiple, small puncta with correspondingly higher cytoplasmic fluorescence (Figure 6, panel A). Strikingly, treatment with 115-7c also produced smaller puncta in Q72- and Q103-expressing cells (Figure 6, panel B). Specifically, cells treated with 115-7c had an average of 5–8 small puncta per cell, whereas mock treated cells typically had 1–2 large puncta (Figure 6, panel C). Importantly, this compound had no effect on Q25- or Q47-expressing cells, suggesting that its effects were specific to aggregation-prone polyQ (Figure 6). Because the potency of 115-7c is modest (~100 μM), we were concerned that this activity might arise from detergent-like or other nonspecific effects. To test this idea, we used SW19, a compound that resembles 115-7c but lacks any activity in the ATPase or luciferase-refolding assay (Supplementary Figure 4A–C). This control compound failed to impact aggregation patterns in cells expressing polyQs (Supplementary Figure 4D,E). Moreover, the inhibitor 116-9e was unable to significantly alter aggregation outcomes in polyQ-expressing cells (Supplementary Figure 4D,E). These results suggest that the stimulatory activity of 115-7c is required for its ability to impact aggregation patterns. In further support of this model, immobilized 115-7c bound to yeast cytosolic Hsp70s (Ssa1p and Ssa2p) in lysates (Supplementary Figure 5A). These results suggest that 115-7c binds to Hsp70 in yeast. However, another possibility is that this molecule stimulates a stress response, because this pathway is also thought to repress polyQ aggregation. To test this idea, we employed LacZ reporters of either the heat shock response or the unfolded protein response. In these systems, 115-7c failed to induce a stress response by either measurement, whereas known positive controls and elevated temperatures both activated the reporters (Supplementary Figure 5B). Finally, 115-7c was relatively stable during the course of these experiments, as measured by LC-MS (Supplementary Figure 6). Together, these studies suggest that the dihydropyrimidine acts through Hsp70s ATPase activity to influence polyQ aggregation.
Figure 6.
Pharmacological stimulation of Hsp70 phenocopies chaperone overexpression in a model of polyQ aggregation. In this model, the length of a polyglutamine domain determines the propensity of the GFP-tagged protein to aggregate into puncta. A) Overexpression of yeast SSA1 or YDJ1 reduces the size of Q103-GFP aggregates and enhances diffuse fluorescence. B) Addition of 100 μM 115-7c reduces the size of the aggregates and enhances the diffuse, background fluorescence in the Q72- and Q103-GFP expressing cells. Scale bars are 3 μm and the arrows indicate puncta. C) Quantification of the results confirms that the number of small aggregates per cell increases after treatment with 115-7c. Error bars represent the standard error of the means from comparing 3–10 random fields.
Conclusions
Dihydropyrimidines, such as 115-7c, have been previously identified as pharmacological modulators of Hsp70, and these molecules were found to display an interesting but unexplained dependence on J-domains for their function (32). To explore the mechanistic origins of this effect, we used a combination of biochemical assays, NMR, and computational modeling to reveal that 115-7c binds to the IIA subdomain and accesses a previously unidentified allosteric pathway. Remarkably, the compound was able to partially suppress ydj1Δ phenotypes in yeast. This stimulatory activity was not competitive with a J-domain co-chaperone, and in fact, these factors appeared to work together to stimulate nucleotide hydrolysis. On the basis of these data, we converted 115-7c into a context-dependent inhibitor, 116-9e, by appending steric bulk. Interestingly, an inhibitor that was previously identified as being sensitive to J-domains, MAL3-101, also has a diphenyl substitution at the same position as 116-9e (32). These results might help clarify how compounds like 115-7c might be reassigned as either activators or inhibitors depending on both their chemical substitution patterns and the presence of J-domains.
Hsp70 is highly conserved among both prokaryotes and eukaryotes, with approximately 50% identity and 70% similarity between E. coli DnaK and other eukaryotic Hsp70s, such as human Hsp72 and yeast Ssa1. Moreover, the binding site for 115-7c, as determined by NMR, is well conserved, with many of the key residues maintained throughout evolution (Supplementary Figure 7). Thus, we hypothesize that 115-7c might act on other Hsp70 isoforms. In support of this idea, 115-7c has activity in yeast (Figure 6). Moreover, it also stimulates human cytoplasmic Hsp70 (33) and related compounds have activity against Ssa1 (32).
An interesting and unexpected aspect of this study is that the small molecules appear to operate at a protein–protein interface to either stimulate or disrupt Hsp70–Hsp40 interactions. A growing number of studies have reported inhibitors of protein–protein interactions (45, 46). In those reports, the molecules typically bind to one protein partner and either directly (47) or allosterically (48, 49) block formation of the protein–protein complex. Some recent examples have even demonstrated that stimulators of protein function can be identified (50, 51), but these compounds are not thought to act at natural protein-protein interfaces. Our findings provide evidence that inhibitory activities at protein–protein contacts are not predestined (52). Together, these findings suggest exciting new avenues for regulating the assembly, disassembly and function of multiprotein complexes (53).
We hypothesize that the unusual mechanism of 115-7c may be an unintended byproduct of the high-throughput screening approach. Specifically, the screen that yielded this compound targeted the combination of DnaK and DnaJ (29). As mentioned above, the J-domain does not have intrinsic ATPase activity but only stimulates the activity of the core Hsp70. Thus, we suspect that screens of the reconstituted complex might have favored identification of a molecule with the observed mechanistic properties at the Hsp70–Hsp40 interface. It is interesting to speculate that additional stimulators and inhibitors might be uncovered using a similar strategy.
More broadly, these findings also establish 115-7c and its derivatives as novel probes to explore Hsp70s important roles in protein homeostasis. Genetic manipulation of Hsp70 is typically not well tolerated, and thus new chemical probes that acutely and transiently disrupt its functions are needed (7, 8). By analogy, inhibitors of Hsp90 have been instrumental in understanding that protein (54). Moreover, a growing number of studies have implicated Hsp70 and its co-chaperones in disease pathways, such as cancer and neurodegeneration (7, 55, 56). In fact, 115-7c has recently been used in cell-based and animal models of tau-based neurodegeneration to reveal that Hsp70 activity is critical for controlling tau stability and quality control (33). In other experiments, this compound was found to influence processing of Akt, another Hsp70 substrate (34), and similar compounds have been found to have anticancer activity (31, 35). In the present study, we found that 115-7c disrupts polyQ aggregation, providing another potential avenue toward pharmacologic intervention. The mechanistic insights provided by this work help clarify why dihydropyrimidines have activity in these disease models, and they suggest an important role for the Hsp70–Hsp40 complex in pathology.
METHODS
Reagents and General Methods
E. coli DnaKFL (residues 1–605), DnaKNBD (residues 1–388), DnaJ and GrpE were expressed in BL21/DE3 and purified as previously described (29). Dihydropyrimidines 115-7c and 116-9e were synthesized by microwave-accelerated Biginelli reactions as described (57). Both the ATPase assays and luciferase refolding assays were also performed as described (29, 30). The concentrations of DnaK DnaJ and GrpE in the ATPase assay were 0.6, 1.0, and 0.9 μM, respectively. The concentrations of DnaK DnaJ and GrpE in the luciferase assay were 0.2, 0.05, and 0.02 μM, respectively.
Yeast Growth Experiments
Wild-type yeast of the W303 background (MATa, leu2-3,112 his3-11 trp1-1 ura3-1 can1-100 ade2-1) and a ydj1Δ strain (DYJ1) (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ydj1∷TRP1) were used. These cells were grown in rich media (YPD) at 30 °C overnight, followed by dilution to OD600 of 0.5. For the experiments performed at elevated temperature, cells were spotted in 5-fold dilutions on YPD agar plates. These plates were pretreated with compound (100 μM) or DMSO alone (0.5%). The plates were grown at 30 or 34 °C for 4 days. In the calcofluor white experiments, the YPD plates were pretreated with this compound to a final concentration of 20 μg mL−1, alone or in combination with 115-7c (100 μM), followed by incubation at 30 °C for 4 days.
NMR Spectroscopy
15N-Labeled DnaKFL(1–605) and DnaKNBD(1–388) were expressed in standard minimal medium containing 15NH4Cl as the sole nitrogen source, using T7-based expression vectors. Following purification, proteins were concentrated to approximately 0.5 mM, exchanged into NMR buffer (25 mM Tris, 10 mM MgCl2, 5 mM KCl, 10% 2H2O, 0.01% sodium azide, pH 7.1), and stored at −80 °C until use. Prior to NMR analysis, DnaK2–388 samples were thawed and supplemented with 5 mM nucleotide (e.g., ATP or ADP) and 10 mM potassium phosphate. Samples of DnaKFL were additionally supplemented with 2.5 mM of a high affinity peptide (NRLLLTG) (41). After collecting reference spectra, 115-7c was added to a final concentration of 1 mM from a 100 mM stock in DMSO. The chemical shifts were in fast exchange on the NMR time scale and were saturable at a 1:1 ratio of compound and DnaK. Control experiments showed that DMSO alone had no detectable effects on the spectrum of DnaKNBD at concentrations up to 1% and minimal effects on the spectra of DnaKFL at the same concentration. 2D HSQC-TROSY NMR spectra were collected at 30 °C on a cryogenic probe-equipped Varian Inova 800 MHz spectrometer, with data collection times of approximately 2 h per spectrum. NMR data were processed using NMRPipe (58) and analyzed with Sparky. Combined 1H and 15N chemical shift changes were measured using a weighted function and the assignments generated previously (41). Residues were selected as significantly affected by the interaction if the combined chemical shift was greater than 0.02 ppm.
Induced Fit Docking (IFD)
The crystal structure of E. coli NBD (PDB: 1DKG) (59) was used after removal of the GrpE fragment and rebuilding of the missing loops using PyMOL (DeLano) and MOE (2005.06). Langevin dynamics simulations of NBD with Amber 10 (60) and FF99SB force field (61) in various nucleotide-binding states were performed to sample relaxed, alternative conformations of the protein. Using snapshots from the simulations, an IFD protocol developed by Schrödinger (2008) was employed (62). Briefly, this procedure involved four core steps: (1) generating ligand poses into the defined binding site by softened-potential docking, (2) refining residue side chains near each ligand pose from the first step, (3) redocking ligand into the optimized induced-fit structure, and (4) scoring the poses by docking energy (GlideScore) and receptor energetic terms (Prime energy). The ligand and protein were prepared in Maestro (2007). IFD was initiated with a constrained minimization of the receptor. The prime loop prediction function of IFD was used to generate a maximum of 5 alternative loop (D208-T215) conformations for Glide docking. Side-chain refinement was performed to residues within 5.0 Å of the defined loop segment. Predicted loop structures with energy 30 kcal/mol above the lowest-energy structure were rejected. In the first stage of docking, Glide docked ligand into the rigid receptor with a softened van der Waals potential. The grid box was centered at residue E206 (based on the NMR results), and the box size was determined automatically. Ten top scoring ligand poses were retained, and for each initial docking pose, Prime performed side-chain refinement to residues that were within 5.0 Å of the ligand, allowing the side chains to adopt optimal conformation to interact with the ligand. Glide then redocked the ligand into the induced-fit receptor with standard parameters, and the final ligand poses were scored using a combination of Glide SP score and Prime energy. Ligand poses with ligand–receptor interactions in agreement with the observed NMR chemical shifts were accepted.
Isothermal Calorimetry
Isothermal calorimetric titrations were performed with a VP-ITC microcalorimeter (MicroCal) at 37 °C. Titration experiments were performed in 25 mM PBS (pH 7.4), 5 mM MgCl2, 150 mM KCl, 5% DMSO. DnaKFL (60 μM), DnaKNBD (60 μM), or DnaJFL (1 mM) were extensively dialyzed and then introduced into the calorimetric cell (cell volume = 1.43 mL). Protein samples were then titrated with 500 μM 115-7c in 15 μL steps (see Supplementary Figure 3 for representative injections). Injections were performed at 2 μL s−1. Data were analyzed using Microcal Origin (v2.9) assuming independent binding sites to yield enthalpy, stoichiometry of binding, and association constants.
Yeast Polyglutamine Aggregation Experiments
Integrated plasmids (p416 vector) expressing huntingtin exon I fragments with 25 and 103 glutamines flanked by an N-terminal Flag tag and a C-terminal green fluorescent protein (GFP) (44) were transformed into a yeast strain of the W303 background. Alternatively, related polyQ constructs were co-expressed in strains harboring galactose-inducible SSA1 (Ssa1 ↑) or YDJ1 (Ydj1 ↑) (Invitrogen). Yeast cultures were grown to midlog phase in selective media (SC-His) for microscopy. If appropriate, the cells were induced by growing in SC-His containing 2% raffinose, followed by centrifugation (2000 rpm, 5 min) and resuspension to an OD600 of 0.3 in SC-His media containing 2% galactose. The cultures were grown at 30 °C in presence of 115-7c (100 μM), 116-9e (100 μM), or an equivalent volume of DMSO (1%). Fluorescence microscopy was performed using a Nikon Eclipse 80i microscope, where the cells were imaged 24 h after induction at 100× magnification. The number of puncta found in cells from 3–10 random fields was used for quantification, and error represents the variance between fields. Nonfluorescent or weakly fluorescent cells were uniformly excluded.
Supplementary Material
Acknowledgment
The authors thank O. Tsodikov, C. White, A. Kumar, and C. Dobry, for helpful conversations; Schrödinger for the generous donation of Glide and Prime software; and D. Klionsky, M. Sherman, S. Lindquist, and R. Sousa for kindly providing strains and plasmids. The authors also acknowledge C. Paulsen and J. Anand for experimental support. This work was supported by the University of Pittsburgh's Center for Chemical Methodologies and Library Development (UPCMLD; P50/GM067082 to P.W.) and the University of Michigan's Community Structural-Activity Resource (CSAR; U01/GM086873 to H.C.), with additional funding from GM75061 (J.L.B.), GM63027 (E.R.P.Z.), NS059690, MCB-0844512 and the McKnight Endowment Fund for Neuroscience (J.E.G.).
Footnotes
Supporting Information Available This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Hohfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001;2:885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pratt WB, Toft DO. Exp. Biol. Med. Vol. 228. Maywood, NJ, U.S.: 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery; pp. 111–133. [DOI] [PubMed] [Google Scholar]
- 6.Nollen EA, Morimoto RI. Chaperoning signaling pathways: molecular chaperones as stress-sensing `heat shock' proteins. J. Cell Sci. 2002;115:2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky JL, Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 2006;6:1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- 8.Patury S, Miyata Y, Gestwicki JE. Pharmacological targeting of the Hsp70 chaperone. Curr. Top. Med. Chem. 2009;9:1337–1351. doi: 10.2174/156802609789895674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buczynski G, Slepenkov SV, Sehorn MG, Witt SN. Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-rate constants. J. Biol. Chem. 2001;276:27231–27236. doi: 10.1074/jbc.M100237200. [DOI] [PubMed] [Google Scholar]
- 10.Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 11.Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- 12.Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in hsp70 chaperones important for allosteric regulation. J. Biol. Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- 13.Pierpaoli EV, Sandmeier E, Schonfeld HJ, Christen P. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J. Biol. Chem. 1998;273:6643–6649. doi: 10.1074/jbc.273.12.6643. [DOI] [PubMed] [Google Scholar]
- 14.Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 2003;42:4937–4944. doi: 10.1021/bi027333o. [DOI] [PubMed] [Google Scholar]
- 16.Siegenthaler RK, Christen P. Tuning of DnaK chaperone action by nonnative protein sensor DnaJ and thermosensor GrpE. J. Biol. Chem. 2006;281:34448–34456. doi: 10.1074/jbc.M606382200. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J co-chaperone binding and regulation of Hsp70. Mol. Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ co-chaperone. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry SJ. Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry. 2003;42:4926–4936. doi: 10.1021/bi027070y. [DOI] [PubMed] [Google Scholar]
- 20.Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA. Interaction of the Hsp70 molecular chaperone, DnaK, with its co-chaperone DnaJ. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sell SM, Eisen C, Ang D, Zylicz M, Georgopoulos C. Isolation and characterization of dnaJ null mutants of Escherichia coli. J. Bacteriol. 1990;172:4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, Li SH, Li XJ. Chaperone suppression of cellular toxicity of huntingtin is independent of polyglutamine aggregation. J. Biol. Chem. 2001;276:48417–48424. doi: 10.1074/jbc.M104140200. [DOI] [PubMed] [Google Scholar]
- 24.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum. Mol. Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 25.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Braun AP, Braun JE. Biological roles of neural J proteins. Cell. Mol. Life Sci. 2008;65:2385–2396. doi: 10.1007/s00018-008-8089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brodsky JL. Selectivity of the molecular chaperone-specific immunosuppressive agent 15-deoxyspergualin: modulation of Hsc70 ATPase activity without compromising DnaJ chaperone interactions. Biochem. Pharmacol. 1999;57:877–880. doi: 10.1016/s0006-2952(98)00376-1. [DOI] [PubMed] [Google Scholar]
- 29.Chang L, Bertelsen EB, Wisén S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal. Biochem. 2008;372:167–176. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Wisen S, Gestwicki JE. Identification of small molecules that modify the protein folding activity of heat shock protein 70. Anal. Biochem. 2008;374:371–377. doi: 10.1016/j.ab.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Wright CM, Chovatiya RJ, Jameson NE, Turner DM, Zhu G, Werner S, Huryn DM, Pipas JM, Day BW, Wipf P, Brodsky JL. Pyrimidinone-peptoid hybrid molecules with distinct effects on molecular chaperone function and cell proliferation. Bioorg. Med. Chem. 2008;16:3291–3301. doi: 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fewell SW, Smith CM, Lyon MA, Dumitrescu TP, Wipf P, Day BW, Brodsky JL. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J. Biol. Chem. 2004;279:51131–51140. doi: 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- 33.Jinwal UK, Miyata Y, Koren J, Jones JR, Trotter JH, Chang L, O'Leary JM, Morgan D, Lee DC, Shults CL, Rousaki A, Weeber EJ, Zuiderweg ER, Gestwicki JE, Dickey CA. Chemical manipulation of Hsp70 reveals its role in tau degradation. 2009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koren J, 3rd, Jinwal UK, Jin Y, O'Leary J, Jones JR, Johnson AG, Blair LJ, Abisambra JF, Chang L, Miyata Y, Cheng AM, Guo J, Cheng JQ, Gestwicki JE, Dickey CA. Facilitating Akt clearance via manipulation of Hsp70 activity and levels. J. Biol. Chem. 2010;285:2498–2505. doi: 10.1074/jbc.M109.057208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodina A, Vilenchik M, Moulick K, Aguirre J, Kim J, Chiang A, Litz J, Clement CC, Kang Y, She Y, Wu N, Felts S, Wipf P, Massague J, Jiang X, Brodsky JL, Krystal GW, Chiosis G. Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat. Chem. Biol. 2007;3:498–507. doi: 10.1038/nchembio.2007.10. [DOI] [PubMed] [Google Scholar]
- 36.Galam L, Hadden MK, Ma Z, Ye QZ, Yun BG, Blagg BS, Matts RL. High-throughput assay for the identification of Hsp90 inhibitors based on Hsp90-dependent refolding of firefly luciferase. Bioorg. Med. Chem. 2007;15:1939–1946. doi: 10.1016/j.bmc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy EJ, McCarty J, Bukau B, Chirico WJ. Conserved ATPase and luciferase refolding activities between bacteria and yeast Hsp70 chaperones and modulators. FEBS Lett. 1995;368:435–440. doi: 10.1016/0014-5793(95)00704-d. [DOI] [PubMed] [Google Scholar]
- 38.Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homo-logues of Escherichia coli dnaJ: a diverse protein family that functions with hsp70 stress proteins. Mol. Biol. Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright CM, Fewell SW, Sullivan ML, Pipas JM, Watkins SC, Brodsky JL. The Hsp40 molecular chaperone Ydj1p, along with the protein kinase C pathway, affects cell-wall integrity in the yeast Saccharomyces cerevisiae. Genetics. 2007;175:1649–1664. doi: 10.1534/genetics.106.066274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revington M, Zhang Y, Yip GN, Kurochkin AV, Zuiderweg ER. NMR investigations of allosteric processes in a two-domain Thermus thermophilus Hsp70 molecular chaperone. J. Mol. Biol. 2005;349:163–183. doi: 10.1016/j.jmb.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 41.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the inter-domain linker. Mol. Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Awad W, Estrada I, Shen Y, Hendershot LM. BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1164–1169. doi: 10.1073/pnas.0702132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 46.Berg T. Modulation of protein-protein interactions with small organic molecules. Angew. Chem., Int. Ed. 2003;42:2462–2481. doi: 10.1002/anie.200200558. [DOI] [PubMed] [Google Scholar]
- 47.Arkin MR, Randal M, DeLano WL, Hyde J, Luong TN, Oslob JD, Raphael DR, Taylor L, Wang J, McDowell RS, Wells JA, Braisted AC. Binding of small molecules to an adaptive protein-protein interface. Proc. Natl. Acad. Sci. U.S.A. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorczynski MJ, Grembecka J, Zhou Y, Kong Y, Roudaia L, Douvas MG, Newman M, Bielnicka I, Baber G, Corpora T, Shi J, Sridharan M, Lilien R, Donald BR, Speck NA, Brown ML, Bushweller JH. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chem. Biol. 2007;14:1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 50.Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Putt KS, Chen GW, Pearson JM, Sandhorst JS, Hoagland MS, Kwon JT, Hwang SK, Jin H, Churchwell MI, Cho MH, Doerge DR, Helferich WG, Hergenrother PJ. Small-molecule activation of procaspase-3 to caspase-3 as a personalized anticancer strategy. Nat. Chem. Biol. 2006;2:543–550. doi: 10.1038/nchembio814. [DOI] [PubMed] [Google Scholar]
- 52.Gestwicki JE, Marinec PS. Chemical control over protein-protein interactions: Beyond inhibitors. Comb. Chem. High Throughput Screening. 2007;10:667–675. doi: 10.2174/138620707782507296. [DOI] [PubMed] [Google Scholar]
- 53.Gordo S, Giralt E. Knitting and untying the protein network: modulation of protein ensembles as a therapeutic strategy. Protein Sci. 2009;18:481–493. doi: 10.1002/pro.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neckers L. Chaperoning oncogenes: Hsp90 as a target of geldanamycin. Handb. Exp. Pharmacol. 2006:259–277. doi: 10.1007/3-540-29717-0_11. [DOI] [PubMed] [Google Scholar]
- 55.Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007;581:3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- 56.Barral JM, Broadley SA, Schaffar G, Hartl FU. Roles of molecular chaperones in protein misfolding diseases. Semin. Cell Dev. Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Wisen S, Androsavich J, Evans CG, Chang L, Gestwicki JE. Chemical modulators of heat shock protein 70 (Hsp70) by sequential, microwave-accelerated reactions on solid phase. Bioorg. Med. Chem. Lett. 2008;18:60–65. doi: 10.1016/j.bmcl.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 58.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 59.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 60.Walker RC, Crowley MF, Case DA. The implementation of a fast and accurate QM/MM potential method in Amber. J. Comput. Chem. 2008;29:1019–1031. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- 61.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.