Figure 1. Microduplications and microdeletions at 16p11.2 in persons with schizophrenia and controls.
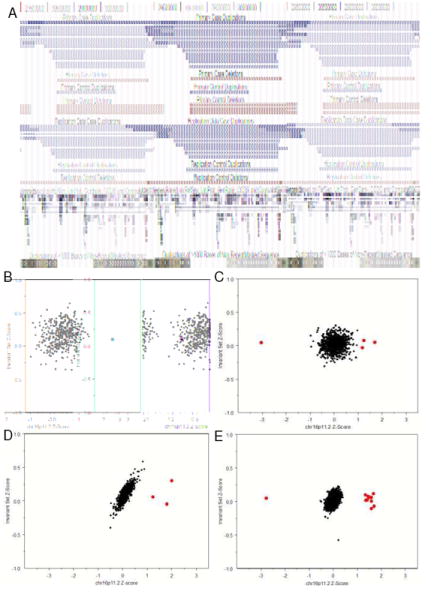
(A) 16p11.2 rearrangements were detected in a primary sample of 1906 cases and 3971 controls (Panels A, B, C, D) and a replication sample of 2645 cases and 2420 controls (Panel A, E). The single microduplication and three microdeletions detected in the primary control set are presented based on the Affymetrix 500K coordinates (hg18). All other CNVs were validated in the NimbleGen HD2 platform and are illustrated based on the validation coordinates (Panels B, C, D, E) The median z-score for the 535kb 16p11.2 target region is plotted on the X-axis and the median z-score of flanking invariant probes is plotted on the Y-axis. Data are presented separately for the ROMA (B), Affymetrix500K (C), NimbleGen HD2 (D), and (Affymetrix 6.0 (E) platforms. CNVs were called using thresholds of >2 SD for ROMA and >1 SD for all other platforms ( ). MeZOD and the HMM algorithms detected the same deletions and duplications at 16p11.2.
). MeZOD and the HMM algorithms detected the same deletions and duplications at 16p11.2.
