FIGURE 1.
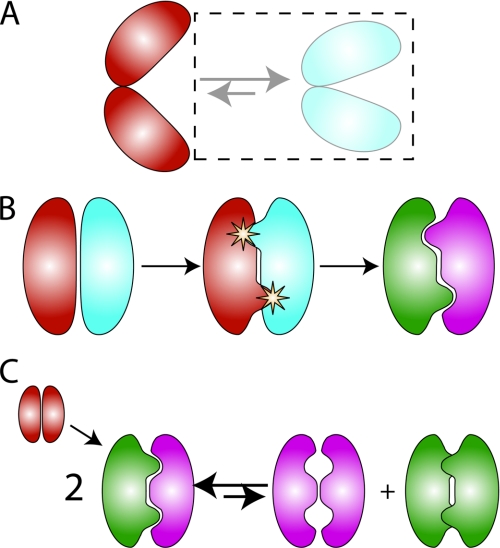
Computational strategies for achieving specificity. A, positive design only. When the desired and undesired states are structurally distinct, as shown for hypothetical open and closed states for a protein, positive design may be sufficient, and negative design states may be ignored (dashed box on the right side of the equilibrium). B, second-site suppressor strategy for redesigning protein-protein interfaces. Negative and positive design elements may be added sequentially. Here, the design proceeds in two steps. First, point mutations are identified on both partners in the interaction that destabilize the native complex. Second, compensatory mutations are identified that restore affinity while accommodating the specificity-conferring amino acids. C, explicit multistate design. Here, a symmetric homodimer (shown in red) is converted into an obligate heterodimer (green and purple). The protein sequence is simultaneously selected both to stabilize the heterodimeric positive design state (left side of the equilibrium) and to destabilize the homodimeric negative design states.