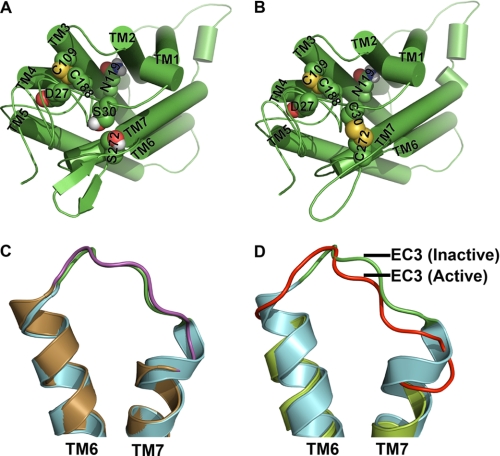
FIGURE 11.
Hypothetical molecular models of C5aR and illustration of conformational changes in EC3 due to disulfide linkage and receptor activation. Molecular models of wild type and engineered C5aR are presented, respectively, in A and B. Asp27, Ser30, Asn119, and Ser272 that have been mutated for engineering the C5aR are highlighted in spheres. B, highlight of both the conserved and the engineered disulfide bond, respectively, between Cys109–Cys188 and Cys30–Cys272 in spheres, in context of other residues. Loop structures have been smoothed for clarity. C, structural alignment of EC3 of wild type (green loop with cyan helices, 1F88) and engineered rhodopsin (magenta loop with sand helices, Protein Data Bank code 2J4Y). Disulfide linkage between Asp282 of EC3 with Asn2 of the N terminus increases the EC3 loop length by straightening the TM7 by one helical loop. D, structural alignment of EC3 of rhodopsin (green loop with cyan helices, Protein Data Bank code 1F88) with EC3 of apparently active opsin (red loop with lime helices, Protein Data Bank code 3DQB). Major conformational changes noted in EC3 are associated with straightening of both TM6 and TM7 helices.