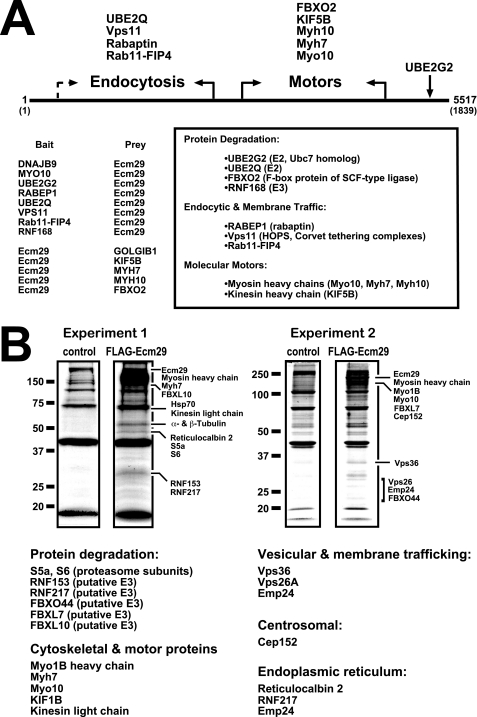
FIGURE 1.
Ecm29 interacting proteins. A, identification of Ecm29-binding proteins by yeast two-hybrid screens. The cDNA libraries used in the yeast two-hybrid screens were collections of fragmented cDNAs encoding on average, 200–300 amino acids (supplemental Table 3). Thus, it was possible to place binding sites for endocytic components in the N-terminal half of Ecm29 and for motor proteins in the C-terminal portion of the protein (see “Experimental Procedures” for details). When both bait and prey Ecm29 cDNAs were isolated for a given class of proteins, the functional boundary is indicated with solid arrows. The dashed arrow indicates an instance in which a functional boundary could not be firmly defined on the basis of available sequences (supplemental Table 3). B, identification of Ecm29-binding proteins by mass spectrometry. HEK293 cells expressing FLAG-Ecm29 were lysed in buffer containing Triton X-100, and the post-mitochondrial supernatants were incubated with M2TM anti-FLAG agarose beads. Bound proteins were eluted with 3× FLAG peptide. As controls, extracts from non-transfected cells were prepared and incubated with antibody beads. Samples (60 μl) of the FLAG-peptide-released proteins were separated by SDS-PAGE and stained with Colloidal Coomassie® Blue. Protein bands in the FLAG sample that were not present in the control lane were excised and subjected to in situ trypsin digestion followed by LC-MS/MS for identification. Corresponding regions in the control lanes were also excised and subjected to LC-MS/MS for comparison. Note that all of the listed Ecm29 interactors were absent in the control samples. A third experiment (not shown) confirmed the results presented above.