Abstract
Type I transglutaminase (TG1) is an enzyme that is responsible for assembly of the keratinocyte cornified envelope. Although TG1 mutation is an underlying cause of autosomal recessive congenital ichthyosis, a debilitating skin disease, the pathogenic mechanism is not completely understood. In the present study we show that TG1 is an endoplasmic reticulum (ER) membrane-associated protein that is trafficked through the ER for ultimate delivery to the plasma membrane. Mutation severely attenuates this processing and a catalytically inactive point mutant, TG1-FLAG(C377A), accumulates in the endoplasmic reticulum and in aggresome-like structures where it is ubiquitinylated. This accumulation results from protein misfolding, as treatment with a chemical chaperone permits it to exit the endoplasmic reticulum and travel to the plasma membrane. ER accumulation is also observed for ichthyosis-associated TG1 mutants. Our findings suggest that misfolding of TG1 mutants leads to ubiquitinylation and accumulation in the ER and aggresomes, and that abnormal intracellular processing of TG1 mutants may be an underlying cause of ichthyosis.
Keywords: Apoptosis, Cell Differentiation, Differentiation, Endoplasmic Reticulum (ER), Intracellular Trafficking, ER Stress, Keratinocyte, Aggresome, Epidermis, Transglutaminase
Introduction
Transglutaminases comprise a family of multifunctional proteins that play an important role in protein stabilization and intracellular signaling (1, 2). The consensus view is that epidermal type I transglutaminase (TG1),2 which is expressed in surface epithelia, has an important and essential role in catalyzing protein-protein cross-link formation leading to formation of the cornified envelope (3–8). The cornified envelope is a 15-nm thick structure comprised of covalently cross-linked proteins and lipids deposited adjacent to the inner surface of the plasma membrane in differentiating keratinocytes (7, 9–16). It is assembled from soluble (e.g. involucrin and small proline-rich proteins) and non-soluble (e.g. loricrin, periplakin, and envoplakin) proteins (17, 18). TG1 catalyzes the formation of protein-protein bonds in which the amine acceptor is provided by the ϵ-amino group of a protein-bound lysine and the ultimate link is a N6-(γ-glutamyl)lysine isopeptide bond (19, 20). The cornified envelope is an essential component of the epidermal barrier. Indeed a key role for TG1 in barrier assembly is indicated by impaired barrier function in Tgm1 knock-out mice (21). TG1 function is also required for normal epidermal function in humans. TG1 mutations are present in 50% of autosomal recessive congenital ichthyosis patients. Autosomal recessive congenital ichthyosis is a debilitating skin disease characterized by scaly epidermis and reduced barrier function. Over 90 different mutations of the Tgm1 gene have been identified in these patients (22). Many of these mutations are deletion or point mutations within the catalytic domain, but disease-associated mutations are also located in other segments of the TG1 protein that do not include residues that are directly required for activity (22, 23). These mutations are associated with reduced TG1 level and activity in tissue and cultured cells derived from patients (24).
We know little about how TG1 is trafficked within cells and the impact of disease-associated mutation on these processes. In the present report we study TG1 trafficking and the impact of TG1 mutation on this process and on cell phenotype. Our studies indicate that TG1 is trafficked and processed in the ER and then delivered to the plasma membrane. In contrast, ichthyosis-associated TG1 mutants accumulate in the endoplasmic reticulum and are ubiquitinylated and also shuttled to aggresomes. We propose that inappropriate accumulation of mutant TG1 in intracellular organelles is a potential underlying cause of autosomal recessive congenital ichthyosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Adenovirus Infection and Plasmid Transfection
Primary cultures of human foreskin keratinocytes were cultured in 0.09 mm calcium-containing keratinocyte serum-free medium (25, 26). For stratified air/liquid interface cultures, two million keratinocytes were plated on a Millicell PCF membrane (0.4 μm, 12-mm insert, 0.6-cm2 surface area) and maintained in Epilife medium supplemented with 1.5 mm calcium for 4 days prior to harvest. Construction of tAd5-TG1-FLAG was previously described (27). tAd5-EV is an empty adenovirus (28, 29). The tAd5 adenovirus encodes the tetracycline operator element linked to the cytomegalovirus promoter. This promoter is active in the presence of the tetracycline-bound transactivator protein, which is provided by co-infection with an Ad5-transactivator adenovirus (28, 29). For experiments, keratinocytes were incubated with 2.5–10 m.o.i. of tAd5-EV or tAd5-TG1-FLAG in the presence of 5 m.o.i. of Ad5-transactivator in keratinocyte serum-free medium containing 6 μg/ml of Polybrene (Sigma). pcDNA3-TG1-FLAG(R142C) and pcDNA3-TG1-FLAG(V379L) were produced by site-directed mutagenesis to convert R142C and V379L. pcDNA3-TG1-FLAG(C377A) and pΔE1Sp1Btet-TG1-FLAG(C377A) were produced by site-directed mutagenesis to convert cysteine 377 of TG1 to alanine. pΔE1Sp1Btet-TG1-FLAG(C377A) was then packaged with the pJM17 adenovirus backbone to produce the tAd5-TG1-FLAG(C377A) adenovirus. PCR primers were used to create the TG1 mutant lacking the N-terminal 52 amino acids and having a FLAG epitope at the amino terminus. The PCR product was cloned into EcoRI/XbaI-restricted pcDNA3 to produce pcDNA3-FLAG-TG1(Δ1–52). For plasmid transfection, 60% confluent keratinocytes were transfected with plasmid using FuGENE 6 (Roche Applied Science) (30). Cells were either fixed for immunofluorescence or lysed for immunoblot analysis 24, 48, or 72 h post-transfection.
Immunological Methods
Immunological detection was performed as described previously (25, 26). For immunofluorescence, keratinocytes, grown on coverslips, were infected with adenoviruses, or transfected with plasmids, and at 24, 48, or 72 h post-treatment the cells were washed, fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, and methanol permeabilized for 10 min at −20 °C. The coverslips were then incubated for 1 h each with the appropriate antibodies. After washing, the samples were affixed to slides using Mowiol 4-88 (Calbiochem, San Diego, CA), and fluorescence was visualized using a Zeiss LSM510 confocal, or an Olympus OX81 spinning-disc confocal microscope. For immunoprecipitation, total cell extract (150 μg of protein), prepared in lysis buffer, was pre-cleared by addition of 25 μl of protein A/G-agarose for 1 h at 25 °C. The lysate was then combined with 50 μl of lysis buffer-equilibrated anti-FLAG-agarose and incubated overnight at 4 °C. The anti-FLAG-agarose was then washed thoroughly with lysis buffer, boiled, and the entire lysate was loaded on a polyacrylamide gel followed by detection of co-precipitated proteins by immunoblot.
Rabbit polyclonal anti-type 1 transglutaminase produced against amino acids 731–817 of human TG1 was from Santa Cruz Biotechnology (sc-25786), rabbit polyclonal anti-FLAG (F7425) was from Sigma, and rabbit polyclonal anti-K14 was obtained from Covance (PRB-155P). Murine monoclonal antibodies include anti-TG1 produced against amino acids 2–33 of human TG1 (sc-166467), anti-ubiquitin (sc-8017), and anti-γ-tubulin (sc-17788) from Santa Cruz; anti-BiP (610798), anti-calnexin (610524), and anti-GM130 (610822) were from BD Transduction Laboratory. Anti-FLAG M2 (F1804), peroxidase-conjugated anti-FLAG M2 (A8592), anti-FLAG M2-agarose (A2220), and anti-β-actin (A5441) were obtained from Sigma. Anti-β-tubulin (ab11311-200) was obtained from Abcam. Alexa-conjugated secondary antibodies were purchased from Molecular Probes (A11008, A11029, A21429, A21424, 11046). Peroxidase-conjugated donkey anti-rabbit IgG (NA934) and peroxidase-conjugated sheep anti-mouse IgG (NA931) were purchased from Amersham Biosciences. Fluorescein cadaverine (FC) (5-((5-aminopentyl)thioreidyl)fluorescein) was obtained from Molecular Probes (A10466). Trimethylamine-N-oxide (TMAO) was purchased from Aldrich (317594). Brefeldin A was obtained from Sigma (B5936). Nocodazole was purchased from Calbiochem (486928).
Subcellular Fractionation
Cells were washed with phosphate-buffered saline, harvested by scraping, and collected by centrifugation. Cell pellets were collected on ice and extracted for 10 min in lysis buffer containing 0.25 m sucrose, 10 mm triethanolamine, 1 mm EDTA, 1 mm β-mercaptoethanol, and protease inhibitor mixture, and passed 30 times through a 27.5-gauge needle. The resulting extract was centrifuged at 1,000 × g three times. The final supernatant was centrifuged at 100,000 × g for 1 h to yield microsomes and cytosol. An equal number of cell equivalents of the cytosol and microsome fraction were boiled and electrophoresed for immunoblot. For microsome extraction experiments, identical aliquots of microsome suspension were divided into four tubes and individual tubes were incubated for 30 min on ice with a final concentration of 1 m NaCl, 0.1 m Na2CO3 (pH 11), or 1% Triton X-100. The samples were centrifuged at 100,000 × g for 1 h and the soluble and insoluble fractions were dissolved at 37 °C by addition of SDS to a final concentration of 4%. For proteinase K challenge assay, microsomes were resuspended in protease inhibitor mixture-free lysis buffer. Individual identical aliquots were incubated with 100 μg/ml of Proteinase K with or without 1% of Triton X-100 on ice for 30 min. Phenylmethylsulfonyl fluoride was then added to the resulting fractions prior to immunoblot.
Immunoelectron Microscopy
Keratinocytes were infected with appropriate adenovirus and at 48 h fixed in 0.1 m PIPES buffer (pH 7.4) containing 4% paraformaldehyde. The cells were harvested by gentle scraping, washed with phosphate-buffered saline, pelleted, and embedded in 2.5% low melting temperature agarose. Agarose blocks were trimmed to 1 mm3, washed, and dehydrated by progressively lowering the temperature from 4 to −20 °C and increasing the ethanol concentration. The blocks were infiltrated and embedded in unicryl at −20 °C for 48 h. Ultrathin sections were cut using a Leica UltraE microtome (Leica Microsystems, Inc., Bannockburn, IL) and collected on formvar-coated nickel grids. For immunogold labeling, grids were placed section-side down on a drop of phosphate-buffered saline (pH 7.4) containing 1% BSA, 1% fish gelatin, 0.01 m glycine (blocking solution) for 10 min, and then transferred onto a 10-μl droplet of primary antibody (Santa Cruz, sc25786) diluted in blocking solution and incubated for 30 min at 25 °C. The grids were washed five times in 30 ml of phosphate-buffered saline containing 0.1% BSA and 1% fish gelatin. Antibody binding was visualized using 10-nm gold particle-conjugated goat anti-rabbit IgG (British Biocell International) diluted in blocking solution and incubated and washed as for the primary antibody. Grids were then fixed for 5 min with phosphate-buffered saline containing 2% glutaraldehyde, washed with water, and air dried. Samples were visualized using a Tecnai T12 transmission electron microscope at 80 kV, and images were acquired using an AMT digital camera.
Biochemical Methods
Transglutaminase catalyzed incorporation of [3H]putrescene into dimethylcasein was previously described (31, 32). To monitor activity in cultured cells, keratinocytes, growing on coverslips, were treated with the appropriate adenovirus for 24 h followed by addition of fresh virus-free medium. At 44 h post-infection, fresh keratinocyte serum-free medium, containing 100 μm FC (Molecular Probes, A-10466), was added and the incubation was continued for an additional 4 h (26). To detect disulfide bonds, keratinocytes were infected with tAd5-TG1-FLAG and after 48 h extracts were prepared in phosphate-buffered saline containing 1% Triton X-100. Equivalent quantities of protein were boiled in the presence or absence of dithiothreitol for 5 min and electrophoresed on a 7.5% acrylamide gel lacking reducing agent but containing SDS (33, 34) followed by immunoblot with anti-FLAG.
RESULTS
Intracellular Distribution and Activity of TG1-FLAG
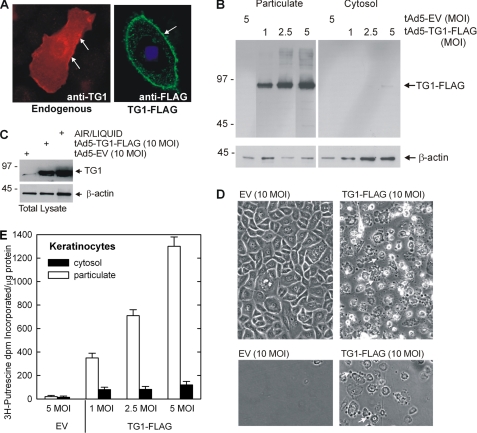
TG1 level and activity are low in undifferentiated cultured normal human epidermal keratinocytes grown in serum-free medium (25, 26, 35), a finding that is consistent with in vivo studies showing that TG1 is absent in undifferentiated cells in epidermis (36–38). Thus, to study TG1 intracellular function we delivered TG1 using a tetracycline-regulated adenovirus, tAd5-TG1-FLAG, that permits expression of full-length TG1-FLAG. Fig. 1A shows that the expressed TG1-FLAG and endogenous TG1 localize to the plasma membrane (arrows), which is consistent with previous observations (39, 40). This association is confirmed by the observation that TG1-FLAG is present in the particulate (membrane) fraction (Fig. 1B). A requirement for these experiments is expression of TG1-FLAG at physiological levels. To demonstrate this, we compared the TG1 level in tAd5-EV and tAd5-TG1-FLAG virus-infected monolayer keratinocytes with the endogenous TG1 level in stratified keratinocyte cultures grown at the air-liquid interface. Air-liquid interface cultures mimic in vivo keratinocyte differentiation (41). The increase observed in raft cultures is consistent with a previous report from Steinert and colleagues (35) that TG1 levels increase more than 100-fold upon keratinocyte differentiation. As shown in Fig. 1C, the TG1 level in tAd5-TG1-FLAG-infected keratinocytes does not exceed the level observed in differentiated air-liquid interface cultures indicating that the expressed TG1 level is in the physiological range. As anticipated, expression of TG1 caused production of cornified envelope-like structures (Fig. 1D, upper panels). These structures display properties of cornified envelopes including resistance to challenge with detergent and reducing agent. Fig. 1D (lower panels) shows survival of cross-linked structures in TG1-FLAG expressing cells after treatment with detergent and reducing agent. To assess activity of expressed TG1-FLAG, particulate and cytosol fractions (Fig. 1B) were assayed for activity. Fig. 1E shows that activity is associated with the particulate fraction and increases with enzyme level. These findings indicate that the expressed TG1-FLAG is active and distributes in the cell in a pattern typical of endogenous TG1.
FIGURE 1.
Expression of TG1-FLAG in keratinocytes. A, keratinocytes growing on coverslips were transfected with 2 μg of pcDNA3 (left) or pcDNA3-TG1-FLAG (right), and after 48 h cells were fixed and stained. Endogenous TG1 (left image, red) and transfected TG1-FLAG (right image, green) are observed at the plasma membrane (arrows) as detected by confocal microscopy (1-μm optical sections). Similar images were observed in each of three experiments. B, TG1-FLAG is membrane associated. Keratinocytes were infected with tAd5-EV or tAd5-TG1-FLAG at the indicated m.o.i. and after 48 h total cell lysates were separated by centrifugation at 15,000 × g into cytosol and particulate fractions. Equal cell equivalents of protein were electrophoresed and TG1-FLAG was detected by anti-FLAG immunoblot (arrow). C, physiological levels of TG1-FLAG. Keratinocytes were grown on at the air/liquid interface for 4 days prior to preparation of extracts for detection of TG1. TG1 expression in the air/liquid interface cultures was compared with that observed in monolayer cultures harvested at 48 h following infection with 10 m.o.i. tAd5-EV and tAd5-TG1-FLAG using anti-TG1 (Santa Cruz, sc-25786). D, TG1-FLAG stimulates cornified envelope formation. Keratinocytes were infected with tAd5-EV or tAd5-TG1-FLAG (top panels). Cells expressing TG1-FLAG undergo marked morphological changes that includes bleb formation (arrows). Dishes washed gently and extracted with Laemmli sample buffer (62.5 mm Tris-HCl, pH 6.8, 5% 2-mercaptoethanol, 2% SDS and 10% glycerol) (bottom panels) reveal insoluble cross-linked structures that remain attached to the dish (75). The arrows indicate the formation of blebs protruding from the cell surface. Approximately 50% of the cells form cornified envelopes under these conditions. E, TG1-FLAG is active. Particulate and cytosol fractions, from the experiment in panel B, were assayed for TG1 activity using the [3H]putrescene incorporation assay. Values are presented as mean ± S.D.
Active-site and Anchor-domain Mutants of TG1
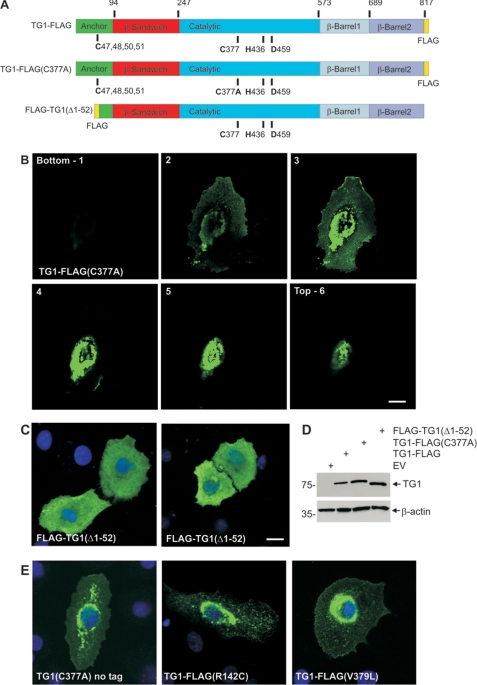
We next assessed the intracellular behavior of several TG1 mutants (Fig. 2A). The TG1 includes membrane-anchoring, β-sandwich, catalytic core, and β-barrel domains (22, 42). Three residues (Cys377, His436, and Asp459) in the TG1 catalytic domain are essential for enzyme activity (43). We first studied a catalytically inactive active-site mutant, TG1-FLAG(C377A). As indicated in the confocal z-series (Fig. 2B), we observe perinuclear accumulation of TG1-FLAG(C377A), although some is also detected at the plasma membrane. This differs from the plasma membrane localization of wild-type TG1-FLAG (Fig. 1A). A second mutant, FLAG-TG1(Δ1–52) lacks the N-terminal 52 amino acids that encode a domain that is thought to be essential for membrane association (40). FLAG-TG1(Δ1–52) localizes to the cytoplasm (Fig. 2C). As shown in Fig. 2D, a difference in expression level does not explain the difference in distribution of these three proteins. We next assessed the localization of two additional TG1 mutants. These point mutations (V379L and R142C) are associated with ichthyosis (23). Val379 is close to the active site and Arg142 is located upstream in the β-sandwich domain. Keratinocytes were transfected with plasmids encoding TG1-FLAG(V379L) and TG1-FLAG(R142C). As shown in Fig. 2E, these mutants distribute in a perinuclear location similar to that observed for TG1(C377A). In addition, to exclude the possibility that the FLAG epitope has an impact on distribution, we monitored the distribution of the C377A mutant lacking the FLAG sequence. As shown in Fig. 2E, the distribution of TG1(C377A) is identical to that observed for TG1-FLAG(C377A) (Fig. 2B).
FIGURE 2.
Subcellular location of TG1 mutants. A, structure of TG1 showing the anchor, β-sandwich, catalytic, and β-barrel 1 and β-barrel 2 domains and the three residues that comprise the catalytic triad (Cys377, His435, and Asp459) and the four (C47,48,50,51) of the five cysteines that form the site that is lipid modified to add the membrane anchor. The position of the FLAG epitope tag is indicated. The numbers at the top indicate amino acids residue. The site of the mutation in TG1-FLAG(C377A) is indicated. FLAG-TG1(Δ1–52) is a truncation mutant. B, subcellular distribution of TG1-FLAG(C377A). Keratinocytes on coverslips were transfected with 2 μg of pcDNA3-TG1-FLAG(C377A) and after 48 h fixed and stained with anti-FLAG (green). The z-stack of images was generated with the bottom picture being at the coverslip/cell junction. C, cells were transfected with 2 μg of pcDNA3-FLAG-TG1(Δ1–52) and after 48 h fixed and stained with anti-FLAG. Nuclei were visualized using Hoescht stain. The bar = 10 μm in all panels. D, to monitor TG1-FLAG, TG1-FLAG(C377A) and FLAG-TG1(Δ1–52) expression cells were transfected with 10 μg of the indicated plasmid and after 48 h extracts were prepared for anti-FLAG immunoblot. E, cells were transfected with plasmids encoding TG1(C377A)(no tag), TG1-FLAG(R142C), and TG1-FLAG(V379L) and stained at 24 h with anti-TG1 (left panel) or anti-FLAG (middle and right panels). Nuclei were visualized using Hoescht stain. All images are 1-μm confocal optical sections.
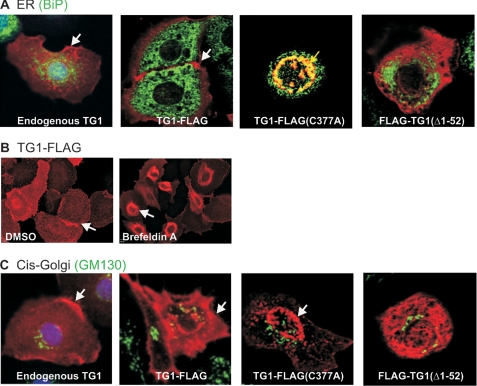
Little is known regarding the mechanism of TG1 intracellular trafficking except that it is proposed to shuttle between the cytoplasm and plasma membrane dependent upon the presence of myristoyl or palmitoyl lipids attached to the N-terminal anchor domain (35, 44). Because TG1 localizes to the plasma membrane (2, 45), we explored the possibility that it may be trafficked via the ER. Fig. 3A shows that endogenous TG1 and TG1-FLAG distribute to the cell membrane (red, arrows) and, in particular, to sites of cell-cell contact, and that FLAG-TG1(Δ1–52) localizes in the cytoplasm. However, these proteins do not localize with BiP (green), a rough ER protein (Fig. 3A). BiP (binding immunoglobulin protein) is a resident protein of the ER lumen that acts as a chaperone to facilitate correct protein folding (46). It is commonly used as an ER marker. Results presented in Fig. 2B indicate that TG1-FLAG(C377A) localizes at a perinuclear location, suggesting it may be associated with the ER. Consistent with ER localization, we show that TG1-FLAG(C377A) colocalizes with BiP (Fig. 3A, yellow staining and arrow) in 40% of TG1-FLAG(C377A) expressing cells, indicating that ER accumulation is a frequent outcome for this mutant (47). In contrast, we observe perinuclear accumulation in only 3.1% of TG1-FLAG expressing cells (not shown), indicating that wild-type TG1 does not accumulate in the ER. To provide evidence that wild-type TG1-FLAG is trafficked via the ER, we treated cells with brefeldin A, an ER transport blocker (48). Brefeldin A treatment results in accumulation of wild-type TG1-FLAG in the ER (Fig. 3A). We also monitored for localization with the cis-Golgi marker, GM130 (49). We did not observe colocalization of endogenous TG1, TG1-FLAG, or TG1 mutants with GM130 (Fig. 3C). Moreover, EGFP-galactotransferase, a marker of the trans-Golgi apparatus, does not colocalize with the wild-type or mutant TG1 (not shown). These findings suggest that wild-type TG1 is rapidly trafficked through the ER but that TG1-FLAG(C377A) accumulates in the ER.
FIGURE 3.
TG1-FLAG intracellular localization. Keratinocytes were transfected with 2 μg of plasmid encoding the indicated proteins and at 48 h cells were stained with the appropriate antibody. A, TG1-FLAG(C377A) colocalizes with BiP. Cells were fixed and stained with anti-FLAG (right three images, red) or rabbit anti-TG1 (left image, red) and with anti-BiP (green). The white arrows indicate TG1 (left panel) and TG1-FLAG location (second from left panel). The yellow arrow (third from left panel) indicates the perinuclear TG1-FLAG(C377) colocalization with BiP. These are confocal 1-μm optical sections. B, TG1-FLAG accumulates in ER with brefeldin A treatment. Keratinocytes were infected with 10 m.o.i. of tAd5-TG1-FLAG and after 3 h treated with 10 μm brefeldin A for 18 h. The cells were stained with anti-FLAG for epifluorescence detection (arrows). C, wild-type and TG1 mutant proteins do not localize with GM130. Cells were incubated with anti-FLAG (right three images) or anti-TG1 (left image) (red) and with anti-GM130 (green). The white arrows indicate TG1 and TG1-FLAG membrane localization, and TG1-FLAG(C377A) perinuclear location. These are confocal 1-μm confocal images.
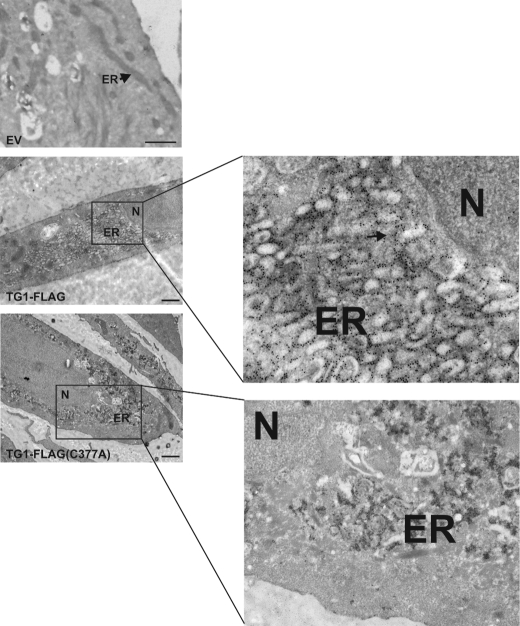
To further pinpoint subcellular location, TG1-FLAG and TG1-FLAG(C377A) were expressed in keratinocytes and distribution was monitored by immunogold electron microscopy. When expressed at physiological levels TG1-FLAG localizes with the ER in a small percentage (3.1%) of cells and these rare cells cannot readily be located in the EM. When expressed at identical levels, TG1-FLAG(C377A) accumulates in the ER in the majority of cells and causes ER swelling (Fig. 4). Such swelling is not observed when TG1-FLAG is expressed at physiological levels, but is observed when TG1-FLAG is expressed at five times higher levels (Fig. 4). As expected, no signal and no morphological changes were observed in EV-infected cells (Fig. 4). These findings demonstrate that TG1-FLAG(C377A), when expressed at physiological levels, tends to accumulate in the ER, but TG1-FLAG does not.
FIGURE 4.
EM detection of TG1-FLAG and TG1-FLAG(C377A). Keratinocytes were infected with empty virus (EV) or virus encoding TG1-FLAG or TG1-FLAG(C377A) and then processed for transmission electron microscopy. The ER and nucleus (N) are indicated. The arrows in the enlarged panels indicate clusters of colloidal gold particles localized in the ER. Bars = 500 nm.
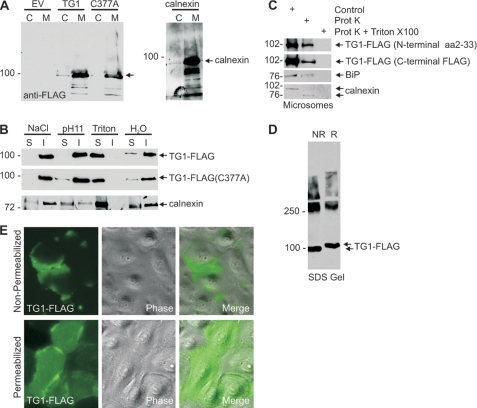
We next used biochemical methods to examine TG1-FLAG and TG1-FLAG(C377A) subcellular distribution. Microsome and cytosol fractions were prepared by 100,000 × g centrifugation. As shown in Fig. 5A, TG1-FLAG and TG1-FLAG(C377A) are enriched in the microsomal (M) fraction, as is the ER marker, calnexin (50). To characterize TG1-FLAG membrane association, the microsomal suspension was extracted with 1 m NaCl, which extracts peripheral membrane proteins, or elevated pH (0.1 m Na2CO3, pH 11), which releases intraluminal proteins (51). As shown in Fig. 5B, TG1-FLAG is not extracted by high salt (NaCl) or elevated pH (pH 11), but is extracted by treatment with Triton X-100, which extracts membrane proteins. We next assessed whether TG1-FLAG is facing the ER lumen. Proteins that are inside the ER are protected from proteinase K digestion. We show that BiP, an ER lumen protein (52, 53), and calnexin, a resident ER transmembrane protein (50, 54), are partially protected from proteinase digestion (Fig. 5C). Calnexin is an ER protein that spans the ER membrane and is therefore reduced in size following proteinase K digestion (50, 54). TG1-FLAG is also partially protected when monitored using antibodies that detect either the N or C terminus. The fact that protection of these proteins is only partial is due to partial loss of membrane integrity during microsome preparation. As expected, all proteins are digested when the microsomal membranes are solublized with Triton X-100 prior to proteinase K treatment. To provide additional evidence that TG1 is processed via the ER, we tested for the presence of intramolecular disulfide bonds in the TG1 protein. Protein lysates were prepared in reducing agent-free conditions and aliquots of lysate were then treated or not treated with reducing agent and electrophoresed on a reducing agent-free SDS gel. As shown in Fig. 5D, reducing agent-treated TG1-FLAG migrates more slowly than non-reduced TG1-FLAG suggesting that TG1-FLAG contains disulfide bonds. Disfulfide bond formation is a post-translational modification that occurs exclusively in the ER (55). Staining of permeabilized cells shows that TG1-FLAG is present on the cell interior (Fig. 5E), but it is also detected in non-permeabilized cells, which suggests it is also on the cell exterior. In both cases, it appears to be present at high levels at points of cell-cell contact. Finding TG1-FLAG outside the cell is consistent with it being in the ER, as ER proteins are often delivered to the cell exterior. Taken together, these findings strongly suggest that TG1 is processed through the ER.
FIGURE 5.
TG1 is associated with the ER membrane. A, microsomal localization of TG1-FLAG and TG1-FLAG(C377A). Keratinocytes were infected with 10 m.o.i. of tAd5-EV, tAd5-TG1-FLAG, or tAd5-TG1-FLAG(C377A) and at 48 h total cell lysates were prepared and separated into cytosol (C) and 100,000 × g pellet (microsomal, M) fractions. Equal cell equivalents of each fraction were electrophoresed for detection of anti-FLAG and anti-calnexin. The arrow indicates migration of TG1-FLAG and TG1-FLAG(C377A). B, TG1-FLAG is associated with the ER membrane. Microsomal membranes were extracted on ice for 30 min with 1 m NaCl, 0.1 m Na2CO3 (pH 11), or 1% Triton X-100 followed by centrifugation at 100,000 × g for 1 h. The resulting soluble (S) and insoluble (I) fractions were electrophoresed for immunoblot with anti-FLAG and anti-calnexin. C, TG1-FLAG localizes inside the ER. Microsomes from tAd5-TG1-FLAG-infected cells were divided into identical aliquots and incubated with 100 μg/ml of proteinase K in the absence or presence of 1% of Triton X-100 on ice. After 30 min the samples were electrophoresed for immunoblot with anti-FLAG. D, TG1 disulfide bonds. Lysates from TG1-FLAG expressing cells were prepared in phosphate-buffered saline containing 1% Triton X-100 and boiled for 5 min in the absence (NR) or presence (R) of reducing agent. Extracts were then electrophoresed on a reducing agent-free SDS-containing 7.5% polyacrylamide gel for immunoblot with anti-FLAG. The arrows indicate migration of reduced and non-reduced monomeric TG1-FLAG. The slower migrating material (≥250 kDa) is cross-linked TG1-FLAG. E, intracellular and extracellular TG1-FLAG. At 48 h after infection with 10 m.o.i. of tAd5-TG1-FLAG, cells were fixed with 4% paraformaldehyde with or without methanol permeabilization, and incubated with anti-FLAG. Primary antibody binding was visualized using Alexa 488-conjugated goat anti-rabbit IgG secondary antibody. These are confocal 1-μm images.
TG1-FLAG and TG1-FLAG(C377A)-specific Morphology Change
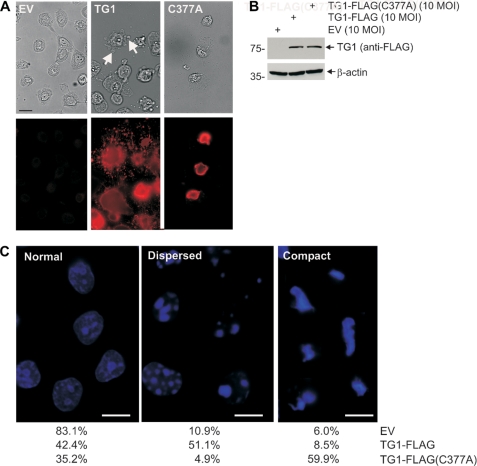
Transglutaminase enzymatic activity is associated with protein-protein covalent cross-link formation (56). Consistent with this idea, expression of physiological levels of active TG1 results in cell morphology change. TG1-FLAG expressing cells display a flattened and fragmented morphology characterized by accumulation of TG1-FLAG positive blebs (Fig. 6A, arrows). In contrast, TG1-FLAG(C377A) expressing cells are reduced in size and are bleb-free (Fig. 6A). Fig. 6B confirms that the morphological differences are not due to a difference in expression level. TG1-FLAG and TG1-FLAG(C377A) also produce distinct changes in nuclear morphology (Fig. 6C). More than 83% of the nuclei in empty vector-infected cells appear “normal.” In contrast, over 50% of cells expressing TG1-FLAG display a loose/punctate chromatin appearance (Fig. 6C, dispersed), and nearly 60% of TG1-FLAG(C377A) expressing cells display a compact chromatin phenotype (Fig. 6C, compact).
FIGURE 6.
TG1-FLAG(C377A) produces a unique phenotype. A, keratinocytes were infected with 10 m.o.i. of tAd5-EV, tAd5-TG1-FLAG, or tAd5-TG1-FLAG(C377A) and at 72 h fixed and stained for morphology assessment and detection of FLAG epitope. The arrows in the TG1 panel identify blebs. B, expression level of TG1 mutants was monitored by immunoblot of total extracts. C, impact on chromatin. Keratinocytes were infected with adenovirus encoding the indicated protein were fixed at 72 h and stained with Hoechst. The percent of cells displaying each nuclear phenotype was determined by counting cells: EV, 265 cells in ten fields; TG1-FLAG, 223 cells in ten fields; TG1-FLAG(C377A), 162 cells in nine fields. Bar = 10 μm. The terms normal, dispersed, and compact are operational descriptions of the chromatin phenotype.
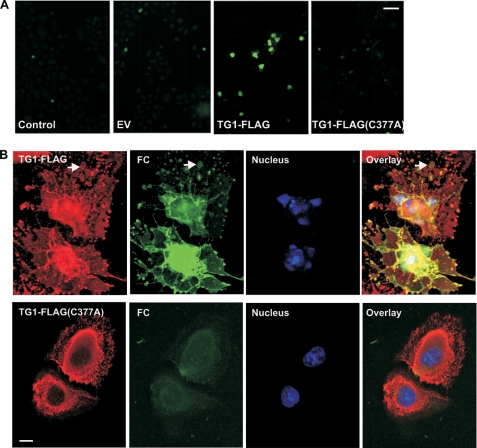
These results suggest that wild-type and TG1-FLAG(C377A) produces different cell changes presumably via different mechanisms. Because TG1-FLAG(C377A) is thought to be catalytically inactive, its effects are presumed not to require cross-linking activity. To confirm that TG1-FLAG(C377A) is inactive in vivo, we expressed TG1-FLAG and TG1-FLAG(C377A) in keratinocytes and assayed for FC incorporation. FC is a cell-permeable fluorescent primary amine (57) that in the presence of transglutaminase activity is covalently attached to protein (58, 59). As shown in Fig. 7A, cells expressing TG1-FLAG display increased transglutaminase activity as monitored by covalent cross-linking of FC (green) to intracellular proteins. In contrast, cells expressing TG1-FLAG(C377A) display background activity equivalent to that observed in empty vector-infected and non-infected (control) keratinocytes. This finding is consistent with our observation that 30–50% of cells infected with 10 m.o.i. of tAd5-TG1-FLAG but no cells infected with 10 m.o.i. of tAd5-TG1-FLAG(C377A) form cornified envelopes (not shown). These findings suggest that the altered cell morphology observed in TG1-FLAG(C377A) expressing cells does not require transglutaminase catalytic activity. Fig. 7B confirms that FC incorporation corresponds to sites of TG1-FLAG localization (e.g. arrows), and that TG1-FLAG(C377A) is not active. Because not all TG1-FLAG expressing cells show FC incorporation, we determined whether the dispersed chromatin phenotype (Fig. 6C, dispersed) requires transglutaminase activity. This analysis reveals that among TG1-FLAG positive cells, 2.5% of FC-negative cells and 67% of FC-positive cells have dispersed chromatin. This suggests that the dispersed chromatin phenotype requires transglutaminase activity.
FIGURE 7.
Transglutaminase activity assay. A, cells were infected with 10 m.o.i. of tAd5-EV, tAd5-TG1-FLAG, or tAd5-TG1-FLAG(C377A) and after 48 h incubated with 100 μm FC for 4 h before detection of FC by epifluorescence. Bar = 50 μm. B, cells, treated as above, were monitored for FC fluorescence (green), TG1-FLAG (anti-FLAG, red), and the nuclear stain (Hoechst, blue). Bar = 10 μm. The signal in the TG1-FLAG(C377A) FC panel is background fluorescence. These are 1-μm confocal sections.
TG1-FLAG(C377A) Accumulates in Aggresomes
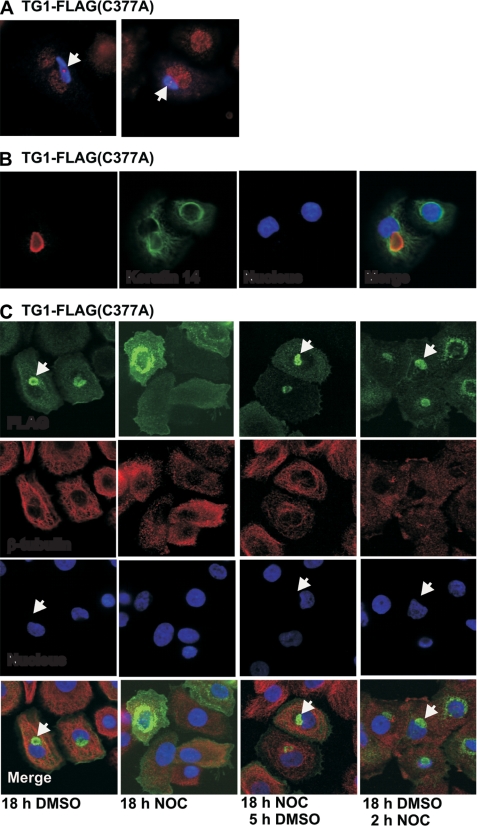
In addition to increased accumulation in the ER, we observed TG1-FLAG(C377A) accumulation in large punctate perinuclear structures in 10% of cells. Such perinuclear structures are characteristic of aggresomes, which colocalize with the centriole/centrosome and contain aggregated misfolded protein. Aggresomes form when cells are unable to remove misfolding proteins via normal proteasome-dependent mechanisms (60–62). We used several criteria to assess whether the perinuclear structures in TG1-FLAG(C377A) expressing cells are aggresomes. First, because aggresomes are known to colocalize with the centrosome, we stained using the centrosome marker, γ-tubulin. As shown in Fig. 8A, TG1-FLAG(C377A) (blue) accumulates in an aggresome-like structure that colocalize with γ-tubulin (red). The intense red γ-tubulin staining in the center of the blue field identifies the centrosome. A second characteristic of aggresomes is that they are surrounded by keratin intermediate filaments (60). Fig. 8B shows the pattern of fluorescence when the keratin 14 intermediate filament protein is stained to identify the intermediate filaments. This shows that the keratin intermediate filaments surround the putative aggresome, in this case formed in response to the presence of the TG1-FLAG(C377A) mutant. A third property is that aggresome formation is microtubule-dependent (60). We therefore examined the effect of treating TG1-FLAG(C377A) expressing cells with the microtubule inhibitor, nocodazole. Keratinocytes were infected with the TG1-FLAG(C377A) encoding virus and at 5 h post-infection cells were treated for 18 h with DMSO, 18 h with nocodazole, 18 h with nocodazole followed by 5 h DMSO, or 18 h with DMSO followed by 2 h with nocodazole. As shown in Fig. 8C, perinuclear structures (arrows) form in DMSO-treated cultures, but not in nocodazole-treated cultures. As anticipated, β-tubulin staining is dispersed in nocodazole-treated cultures indicating the collapse of microtubules. Aggresome formation (arrows) is also observed in 18-h nocodazole-treated cultures when followed by a 5-h washout. This indicates that removing nocodazole leads to aggresome formation. Finally, consistent with the observation that aggresome formation is irreversible, we show that the structures do not disperse when cells are treated with nocodazole for 2 h after being maintained for 18 h in DMSO. Taken together, these results strongly suggest that TG1-FLAG(C377A), and other TG1 mutants (e.g. R142C), accumulate in aggresomes.
FIGURE 8.
TG1-FLAG(C377A) accumulates in aggresomes. A, keratinocytes were transfected with 2 μg of pcDNA3-TG1-FLAG(C377A) and at 48 h fixed and stained with anti-γ-tubulin (red) and anti-FLAG (blue). The arrows identify the centrosome as a point (red) within the blue field. B, keratinocytes were transfected with 2 μg of pcDNA3-TG1-FLAG(C377A) as above, and then co-stained with anti-keratin 14 and anti-FLAG. C, keratinocytes were infected with tAd5-TG1-FLAG(C377A) and after 5 h the cells were treated with 0.1% DMSO for 18 h, 10 μm nocodazole for 18 h, 10 μm nocodazole for 18 h followed by 0.1% DMSO for 5 h, and 0.1% DMSO for 18 h followed by 10 μm nocodazole for 2 h. The cells were stained with anti-FLAG (green) and anti-β-tubulin (red) prior to collection of confocal 1-μm optical sections. The arrows indicate perinuclear aggresome structures.
TG1-FLAG(C377A) Is Ubiquitinated in the Endoplasmic Reticulum and Aggresome
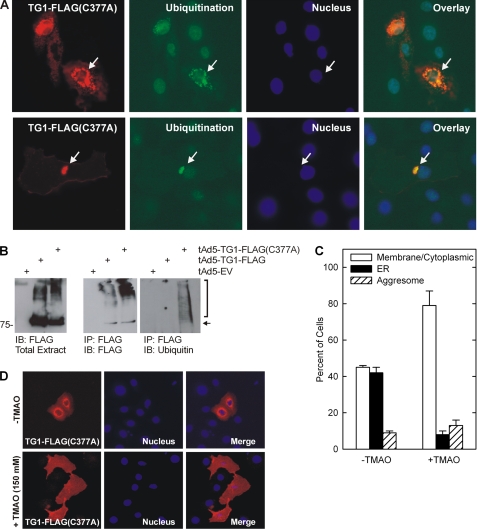
Newly formed proteins are folded in the ER via a molecular chaperone-driven process (47, 63) and proteins that are not properly folded are ubiquitinated and targeted for proteasomal degradation (63). Thus, we examined whether ER-localized TG1-FLAG(C377A) is ubiquitinated. Fig. 9A shows that TG1-FLAG(C377A) colocalizes with ubiquitin in the ER (top panels, arrows) and in aggresome-like perinuclear structures (bottom panels, arrows). Moreover, FLAG epitope precipitation with anti-FLAG followed by immunoblot with anti-ubiquitin reveals that TG1-FLAG(C377A) but not TG1-FLAG is ubiquitinylated (Fig. 9B).
FIGURE 9.
TG1-FLAG(C377A) is ubiquitinylated. A, keratinocytes were transfected with 2 μg of pcDNA3-TG1-FLAG(C377A) and after 48 h stained with anti-ubiquitin (green), anti-FLAG (red), and Hoescht stain. Top and bottom panels show TG1-FLAG(C377A) and ubiquitin colocalization in ER and aggresomes, respectively (arrows). B, TG1-FLAG(C377A) and ubiquitin co-precipitation. Keratinocytes were infected with tAd5-EV, tAd5-TG1-FLAG, or tAd5-TG1-FLAG(C377A) and at 48 h extracts were immunoprecipitated with anti-FLAG (middle and right panels) followed by immunoblot with anti-ubiquitin (right panel) or anti-FLAG (middle panel). Total extract is an immunoblot (IB) of 10% of the amount of protein used for the immunoprecipitation (IP) (left panel). The arrow identifies transglutaminase and the bracket indicates ubiquitinylated TG1-FLAG(C377A). C, TMAO treatment causes TG1-FLAG(C377A) to move from the ER to the plasma membrane. Keratinocytes were transfected as above and after 3 h treated with or without 150 mm TMAO for 48 h and stained with anti-FLAG and the distribution of TG1-FLAG(C377A) was assessed in each of 10 randomly selected fields of 20 cells each. The values are presented as mean ± S.D. This level of TMAO is within the range used in other cell types (65) and did not impact the cells negatively. D, to measure the impact of TMAO on TG1-FLAG(C377A) distribution, TG1-FLAG(C377A) expressing cells were incubated for 3 h with 150 mm TMAO prior to detection with anti-FLAG.
To test the idea that ER accumulation is due to protein misfolding, we employed TMAO, a chemical chaperone that assists protein folding and facilitates movement of misfolded protein out of the ER (64, 65). As shown in Fig. 9C, in the absence of TMAO, 41% of cells display a plasma membrane distribution of TG1-FLAG(C377A), and 44% display ER accumulation. Following TMAO treatment, nearly 80% of cells display a plasma membrane distribution of TG1-FLAG(C377A). Aggresome number remains constant. Fig. 9D shows TG1-FLAG(C377A) redistribution before and following TMAO treatment.
DISCUSSION
Impact of TG1 Expression on Keratinocyte Morphology
Type I transglutaminase is a key regulator of events during terminal keratinocyte differentiation (2, 37, 45, 66–72). It catalyzes the formation of N6-(γ-glutamyl)lysine isopeptide covalent protein-protein bonds that form the cornified envelope (20). Previous studies show that the TG1 level and activity increases in differentiating keratinocytes and that it localizes at the plasma membrane (40, 73). To extend this knowledge, we examined TG1-FLAG function in normal human keratinocytes. We show that TG1-FLAG fractionates with membranes and that expression results in enhanced formation of covalently cross-linked cornified envelope-like structures (i.e. cornified envelopes) that are characterized by resistance to boiling in detergent and reducing agent (4, 74, 75). In addition, TG1-FLAG preferentially localizes at sites of cell-cell contact. This novel observation suggests that TG1 may have a particular role in sealing sites of cell-cell contact, and that mechanisms may exist to guide TG1 to this location. This finding has important implications for understanding the role of TG1 in keratinocytes, because sealing sites of cell-cell contact is important in maintaining barrier competence in vivo. TG1-FLAG expression also produces changes in nuclear chromatin appearance. Unlike normal chromatin, chromatin in TG1-FLAG expressing cells is dispersed at multiple foci, suggesting that it may be fragmented. Nuclear destruction is a property of in vivo differentiating keratinocytes (56) and these findings suggest that TG1 may have a role in promoting this destruction.
TG1 Is Trafficked through the ER
Previous studies indicate that the TG1 N terminus is myristoylated and palmitoylated, that these lipids anchor TG1 to the inner surface of the plasma membrane (39, 40, 44, 73), and that these anchors can be removed resulting in TG1 release into the cytoplasm (39, 40, 44, 73). However, little additional information is available regarding TG1 subcellular distribution and trafficking. We noticed that several TG1 mutants accumulate in the endoplasmic reticulum, suggesting that TG1 is trafficked via the ER. This finding was puzzling, as our initial studies revealed an absence of co-localization of wild-type TG1 with the ER. However, we ultimately confirmed TG1 ER localization using several approaches. First, treatment with the ER protein transport blocker, brefeldin A (48), results in accumulation of TG1 in the ER. Second, expression of TG1-FLAG at supra-physiological levels leads to accumulation in the ER as detected using immunogold electron microscopy. Third, both wild-type and mutant TG1 are protected from digestion when isolated microsomes are challenged with proteinase K, suggesting that TG1 faces the lumenal side of ER. Fourth, we show that TG1-FLAG contains intramolecular disulfide bonds. Such bonds are formed exclusively within the ER by protein disulfide isomerase, an ER-localized enzyme (55, 76). Taken together, these findings suggest that TG1 is processed through the ER. We further propose, based on differential extraction studies, that TG1 is tightly associated with the inner surface of the ER. We do not know how TG1 is trafficked to the ER, as this is thought to require a signal peptide (77, 78) and no such motif is present in the TG1 sequence.
Myristoylation and palmitoylation of the TG1 N terminus has been proposed as required for TG1 plasma membrane association (39, 40, 44, 73). Our present studies confirm that the TG1 N-terminal domain is required for adhesion at the inner surface of the plasma membrane. It can be argued that a tightly associated membrane protein may not require a lipid anchor for membrane association. However, recent studies show that lipid modification can be required for membrane association of integral proteins. An example is cytoskeleton-associated protein 4 (79). Thus, adherence at the inner plasma membrane may require lipid and protein contact with the membrane. In addition, TG1 may associate with the ER membrane via interaction with other proteins. Many key questions remain to be addressed including understanding how ER-trafficked TG1 is transferred to the inner and outer surface of the plasma membrane. We did not find TG1 in the Golgi and so it is possible that processing is rapid or it simply bypasses the Golgi. Bypassing Golgi processing has been reported for other proteins (80). ER-trafficked proteins are known to be released from cells via a membrane fusion process (47, 63). Consistent with a potential extracellular role, it is intriguing that TG1 encodes a RGD tripeptide motif (81), which is characteristic of proteins that interact with fibronectin (81). It will also be important to identify how TG1 is transferred to the cytoplasm to come in contact with enzymes on the cytoplasmic face of the ER and cytoplasm that are involved in myristoylation (82, 83).
Accumulation of TG1-FLAG(C377A) in the ER and Aggresome
TG1 catalytic activity requires a triad of amino acids, including Cys377, His436, and Asp459, which are part of the TG1 catalytic domain (43). It is interesting that the catalytically inactive point mutant, TG1-FLAG(C377A), accumulates in the ER. We propose that TG1-FLAG(C377A) accumulates in the ER because it is misfolded. This idea is supported by the observation that TMAO, a chemical chaperone that facilitates protein folding, causes TG1-FLAG(C377A) to exit the ER. Improperly folded proteins within the ER are ubiquitinated in preparation for removal via a proteasome-dependent mechanism (47, 63, 84). The mutated/misfolded cystic fibrosis transporter, CFTR-ΔF508, for example, is ubiquitinated and degraded via this mechanism (85). Normally, misfolded proteins are released from the ER and delivered to the proteasome for degradation. However, some protein aggregates cannot be processed by the proteasome and, as a result, accumulate in perinuclear structures called aggresomes (60, 86) for subsequent removal by autophagic mechanisms (60, 86). Our studies show that TG1-FLAG(C377A) accumulates in perinuclear structures that we propose are aggresomes because they are compact perinuclear structures; are positive for the centriole-associated protein, γ-tubulin; require microtubule function for formation; and are surrounded by intermediate filaments (61, 62). The presence of TG1 mutant-positive aggresomes suggests that mutant TG1 proteins readily aggregate and are difficult to remove from cells.
ER Accumulation of Misfolded TG1 Mutants: a Mechanism of TG1-related Disease Pathology
Several key observations have been made regarding the role of transglutaminase in ichthyosis. First, minimal changes in the TG1 protein amino acid sequence can cause autosomal recessive congenital ichthyosis (23). For example, 57 of the 94 identified TG1 mutations that cause autosomal recessive congenital ichthyosis are single amino acid changes (22). Second, individual mutants are associated with different disease severity (22–24). Third, most of these patients show reduced levels of TG1 (24). Fourth, on a specific activity basis, most, but not all, of the TG1 mutants that have been examined have reduced activity (87). Fifth, structural analysis suggests that many of these mutations influence protein structure (22–24). This suggests that the mutant TG1 proteins are selectively degraded.
We demonstrate ER accumulation of TG1-FLAG(C377A) in 40% of TG1-FLAG(C377A)-positive cells, and a similar high level of ER accumulation of two ichthyosis-relevant mutants, TG1-FLAG(R142C) and TG1-FLAG(V379L). This is in contrast to ER accumulation of TG1-FLAG in 3% of TG1-FLAG expressing cells. Moreover, this accumulation is associated with ubiquitinylation of the ER-associated mutants. This suggests that the ubiquitin modification system (84) is a key pathway for removing misfolded TG1 protein. Such a sequestration and ubiquitinylation mechanism could explain the ichthyosis-associated reduction in TG1 level.
An additional point to consider is that disease may not be exclusively related to loss of TG1 catalytic activity. We have shown that expressing TG1 mutants produce unique cell phenotypes not observed for normal TG1. For example, expression of TG1-FLAG(C377A) produces a distinct cell and chromatin phenotype despite the fact it is not catalytically active. This suggests that the ichthyosis phenotype may be partially due to cell stress. Finally, it is possible that TG1 levels may be reduced in ichthyosis patients due to accumulation in non-soluble aggregates in aggresomes. Choate et al. (88) have described accumulation of TG1 mutants in congenital ichthyosiform erythroderma.
TG1-FLAG Is Active in Low Calcium Conditions
Our previous studies indicate that TG1 activity is absent in keratinocytes maintained in 0.09 mm calcium-containing (low calcium) medium, but increases in cells growing in medium containing ≥0.2 mm calcium (2, 45). It is interesting that the present experiments show that TG1-FLAG is active when expressed in cells maintained in low (0.09 mm) calcium-containing medium. This is surprising, because calcium is required for transglutaminase activity (89, 90). Our present and past (25, 26) findings suggest that TG1 can be activated without an increase in intracellular calcium. In general the mechanism responsible for this activity is not known, except that it is possible that TG1-interacting proteins have a role (25, 26).
Abnormal TG1 Function in Ichthyosis
In summary, our findings suggest that TG1 is trafficked through the ER in association with membranes and is ultimately exported to the cell exterior or inner surface of the plasma membrane (Fig. 10). In contrast, in ichthyosis, TG1 mutants accumulate in the ER where they are ubiquitinylated. Moreover, this accumulation may be associated with aggregation of the mutant protein in aggresomes. This abnormal trafficking leads to a reduced TG1 level and activity, which are hallmarks of this disease. Moreover, ER-associated TG1 may activate other processes that negatively impact the cell including ER stress (46, 91).
FIGURE 10.
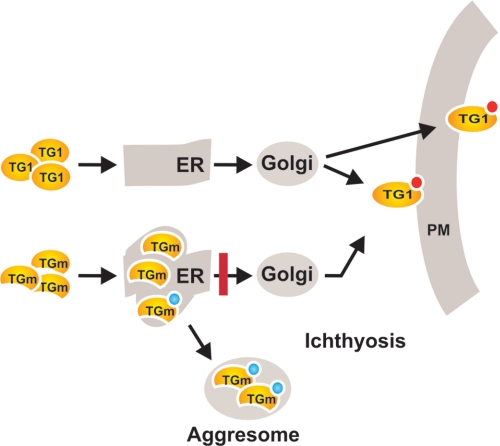
Schematic summary of transglutaminase trafficking. We propose that TG1 is synthesized and trafficked via the ER (and probably the Golgi) for export to the cell exterior and incorporation at the inner surface of the plasma membrane. Our EM and confocal imaging, differential membrane extraction, and proteinase challenge studies suggest that TG1 is associated on the inner surface of the ER lumen. Previous studies show that TG1 is myristoylated and palmitoylated (red circle) and suggest that this is required for TG1 interaction with membranes (39, 40, 44). We are not sure that exported TG1 is myristoylated. In contrast, misfolded TG1 mutants (TGm) are trapped (red bar) in the ER and are ubiquitinylated. Some of this protein is not efficiently removed by proteasome processing and accumulates in aggresomes. We further propose that ER and aggresome accumulation of mutant TG (TGm) contributes to the ichthyosis disease phenotype via two complimentary mechanisms: reducing TG1 level and activity, and activating other cell responses (e.g. ER-associated protein degradation response (63)).
Acknowledgment
Electron microscopy was performed in the University of Maryland Dental School Imaging Core Facility, Director Dr. Ru-ching Hsia.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AR049713 (to R. L. E.).
- TG1
- transglutaminase type 1
- ER
- endoplasmic reticulum
- TMAO
- trimethylamine-N-oxide
- m.o.i.
- multiplicity of infection
- BiP
- binding immunoglobulin protein
- FC
- fluorescein cadaverine
- DMSO
- dimethyl sulfoxide
- EGFP
- enhanced green fluorescent protein
- EV
- empty vector.
REFERENCES
- 1.Lorand L., Graham R. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 140–156 [DOI] [PubMed] [Google Scholar]
- 2.Eckert R. L., Sturniolo M. T., Broome A. M., Ruse M., Rorke E. A. (2005) J. Invest. Dermatol. 124, 481–492 [DOI] [PubMed] [Google Scholar]
- 3.Hennings H., Steinert P., Buxman M. M. (1981) Biochem. Biophys. Res. Commun. 102, 739–745 [DOI] [PubMed] [Google Scholar]
- 4.Robinson N. A., LaCelle P. T., Eckert R. L. (1996) J. Invest. Dermatol. 107, 101–107 [DOI] [PubMed] [Google Scholar]
- 5.Steinert P. M., Candi E., Kartasova T., Marekov L. (1998) J. Struct. Biol. 122, 76–85 [DOI] [PubMed] [Google Scholar]
- 6.Steinert P. M., Marekov L. N. (1997) J. Biol. Chem. 272, 2021–2030 [DOI] [PubMed] [Google Scholar]
- 7.Steven A. C., Steinert P. M. (1994) J. Cell Sci. 107, 693–700 [PubMed] [Google Scholar]
- 8.Yaffe M. B., Murthy S., Eckert R. L. (1993) J Invest. Dermatol. 100, 3–9 [DOI] [PubMed] [Google Scholar]
- 9.Matoltsy A. G. (1976) J. Invest. Dermatol. 67, 20–25 [DOI] [PubMed] [Google Scholar]
- 10.Matoltsy A. G., Odland G. F. (1955) J. Biophys. Biochem. Cytol. 1, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias P. M., Friend D. S. (1975) J. Cell Biol. 65, 180–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grayson S., Elias P. M. (1982) J. Invest. Dermatol. 78, 128–135 [DOI] [PubMed] [Google Scholar]
- 13.Matoltsy A. G., Matoltsy M. N. (1966) J. Invest. Dermatol. 46, 127–129 [PubMed] [Google Scholar]
- 14.Nemes Z., Marekov L. N., Fésüs L., Steinert P. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8402–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinert P. M., Marekov L. N. (1999) Mol. Biol. Cell 10, 4247–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wertz P. W., Swartzendruber D. C., Kitko D. J., Madison K. C., Downing D. T. (1989) J. Invest. Dermatol. 93, 169–172 [DOI] [PubMed] [Google Scholar]
- 17.Eckert R. L., Yaffe M. B., Crish J. F., Murthy S., Rorke E. A., Welter J. F. (1993) J. Invest. Dermatol. 100, 613–617 [DOI] [PubMed] [Google Scholar]
- 18.Kalinin A. E., Kajava A. V., Steinert P. M. (2002) Bioessays 24, 789–800 [DOI] [PubMed] [Google Scholar]
- 19.Folk J. E. (1980) Annu. Rev. Biochem. 49, 517–531 [DOI] [PubMed] [Google Scholar]
- 20.Folk J. E., Finlayson J. S. (1977) Adv. Protein Chem. 31, 1–133 [DOI] [PubMed] [Google Scholar]
- 21.Kuramoto N., Takizawa T., Takizawa T., Matsuki M., Morioka H., Robinson J. M., Yamanishi K. (2002) J. Clin. Invest. 109, 243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farasat S., Wei M. H., Herman M., Liewehr D. J., Steinberg S. M., Bale S. J., Fleckman P., Toro J. R. (2009) J. Med. Genet. 46, 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herman M. L., Farasat S., Steinbach P. J., Wei M. H., Toure O., Fleckman P., Blake P., Bale S. J., Toro J. R. (2009) Hum. Mutat. 30, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber M., Yee V. C., Burri N., Vikerfors E., Lavrijsen A. P., Paller A. S., Hohl D. (1997) J. Biol. Chem. 272, 21018–21026 [DOI] [PubMed] [Google Scholar]
- 25.Sturniolo M. T., Chandraratna R. A., Eckert R. L. (2005) Oncogene 24, 2963–2972 [DOI] [PubMed] [Google Scholar]
- 26.Sturniolo M. T., Dashti S. R., Deucher A., Rorke E. A., Broome A. M., Chandraratna R. A., Keepers T., Eckert R. L. (2003) J. Biol. Chem. 278, 48066–48073 [DOI] [PubMed] [Google Scholar]
- 27.Jans R., Sturniolo M. T., Eckert R. L. (2008) J. Invest. Dermatol. 128, 517–529 [DOI] [PubMed] [Google Scholar]
- 28.Dashti S. R., Efimova T., Eckert R. L. (2001) J. Biol. Chem. 276, 8059–8063 [DOI] [PubMed] [Google Scholar]
- 29.Dashti S. R., Efimova T., Eckert R. L. (2001) J. Biol. Chem. 276, 27214–27220 [DOI] [PubMed] [Google Scholar]
- 30.Efimova T., Broome A. M., Eckert R. L. (2003) J. Biol. Chem. 278, 34277–34285 [DOI] [PubMed] [Google Scholar]
- 31.Kasturi L., Sizemore N., Eckert R. L., Martin K., Rorke E. A. (1993) Exp. Cell Res. 205, 84–90 [DOI] [PubMed] [Google Scholar]
- 32.Sizemore N., Kasturi L., Gorodeski G., Eckert R. L., Jetten A. M., Rorke E. A. (1993) Differentiation 54, 219–225 [DOI] [PubMed] [Google Scholar]
- 33.Braakman I., Hebert D. N. (2001) Curr. Protoc. Protein Sci. 14, 1–15 [DOI] [PubMed] [Google Scholar]
- 34.Liscaljet I. M., Kleizen B., Braakman I. (2004) in Handbood of Protein Folding (Buchner J., Kiefhaber T. eds) Vol. 3, pp. 73–104, Wiley-VCH Verlag GmbH & Co., Weinheim [Google Scholar]
- 35.Steinert P. M., Chung S. I., Kim S. Y. (1996) Biochem. Biophys. Res. Commun. 221, 101–106 [DOI] [PubMed] [Google Scholar]
- 36.Greenberg C. S., Birckbichler P. J., Rice R. H. (1991) FASEB J. 5, 3071–3077 [DOI] [PubMed] [Google Scholar]
- 37.Rice R. H., Chakravarty R., Chen J., O'Callahan W., Rubin A. L. (1988) Adv. Exp. Med. Biol. 231, 51–61 [DOI] [PubMed] [Google Scholar]
- 38.Michel S., Démarchez M. (1988) J. Invest. Dermatol. 90, 472–474 [DOI] [PubMed] [Google Scholar]
- 39.Steinert P. M., Kim S. Y., Chung S. I., Marekov L. N. (1996) J. Biol. Chem. 271, 26242–26250 [DOI] [PubMed] [Google Scholar]
- 40.Phillips M. A., Qin Q., Mehrpouyan M., Rice R. H. (1993) Biochemistry 32, 11057–11063 [DOI] [PubMed] [Google Scholar]
- 41.Poumay Y., Dupont F., Marcoux S., Leclercq-Smekens M., Hérin M., Coquette A. (2004) Arch. Dermatol. Res. 296, 203–211 [DOI] [PubMed] [Google Scholar]
- 42.Kim S. Y., Kim I. G., Chung S. I., Steinert P. M. (1994) J. Biol. Chem. 269, 27979–27986 [PubMed] [Google Scholar]
- 43.Pedersen L. C., Yee V. C., Bishop P. D., Le Trong I., Teller D. C., Stenkamp R. E. (1994) Protein Sci. 3, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravarty R., Rice R. H. (1989) J. Biol. Chem. 264, 625–629 [PubMed] [Google Scholar]
- 45.Eckert R. L., Sturniolo M. T., Broome A. M., Ruse M., Rorke E. A. (2005) Prog. Exp. Tumor Res. 38, 115–124 [DOI] [PubMed] [Google Scholar]
- 46.Ni M., Lee A. S. (2007) FEBS Lett. 581, 3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nebenführ A., Ritzenthaler C., Robinson D. G. (2002) Plant Physiol. 130, 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Short B., Barr F. A. (2003) Curr. Biol. 13, R311–R313 [PubMed] [Google Scholar]
- 50.Bergeron J. J., Brenner M. B., Thomas D. Y., Williams D. B. (1994) Trends Biochem. Sci. 19, 124–128 [DOI] [PubMed] [Google Scholar]
- 51.Ohlendieck K. (2004) Methods Mol. Biol. 244, 283–293 [DOI] [PubMed] [Google Scholar]
- 52.Munro S., Pelham H. R. (1986) Cell 46, 291–300 [DOI] [PubMed] [Google Scholar]
- 53.Bole D. G., Hendershot L. M., Kearney J. F. (1986) J. Cell Biol. 102, 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ou W. J., Bergeron J. J., Li Y., Kang C. Y., Thomas D. Y. (1995) J. Biol. Chem. 270, 18051–18059 [DOI] [PubMed] [Google Scholar]
- 55.Sitia R., Braakman I. (2003) Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 56.Green H. (1980) The Harvey Lectures 74, 101–139 [PubMed] [Google Scholar]
- 57.Pincus J. H., Chung S. I., Chace N. M., Gross M. (1975) Arch. Biochem. Biophys. 169, 724–730 [DOI] [PubMed] [Google Scholar]
- 58.Lajemi M., Demignot S., Borge L., Thenet-Gauci S., Adolphe M. (1997) Histochem. J 29, 593–606 [DOI] [PubMed] [Google Scholar]
- 59.Gray A. C., Clothier R. H. (2001) Toxicol. In Vitro 15, 427–431 [DOI] [PubMed] [Google Scholar]
- 60.Kopito R. R. (2000) Trends Cell Biol. 10, 524–530 [DOI] [PubMed] [Google Scholar]
- 61.Schiebel E. (2000) Curr. Opin. Cell Biol. 12, 113–118 [DOI] [PubMed] [Google Scholar]
- 62.Joshi H. C. (1993) Bioessays 15, 637–643 [DOI] [PubMed] [Google Scholar]
- 63.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 64.Zou Q., Bennion B. J., Daggett V., Murphy K. P. (2002) J. Am. Chem. Soc. 124, 1192–1202 [DOI] [PubMed] [Google Scholar]
- 65.Gong B., Zhang L. Y., Pang C. P., Lam D. S., Yam G. H. (2009) Mol. Vis. 15, 2829–2840 [PMC free article] [PubMed] [Google Scholar]
- 66.Ogawa H., Goldsmith L. A. (1976) J. Biol. Chem. 251, 7281–7288 [PubMed] [Google Scholar]
- 67.Negi M., Colbert M. C., Goldsmith L. A. (1985) J. Invest. Dermatol. 85, 75–78 [DOI] [PubMed] [Google Scholar]
- 68.Steinert P. M., Candi E., Tarcsa E., Marekov L. N., Sette M., Paci M., Ciani B., Guerrieri P., Melino G. (1999) Cell Death. Differ. 6, 916–930 [DOI] [PubMed] [Google Scholar]
- 69.Russell L. J., DiGiovanna J. J., Rogers G. R., Steinert P. M., Hashem N., Compton J. G., Bale S. J. (1995) Nat. Genet. 9, 279–283 [DOI] [PubMed] [Google Scholar]
- 70.Kim S. Y., Chung S. I., Steinert P. M. (1995) J. Biol. Chem. 270, 18026–18035 [DOI] [PubMed] [Google Scholar]
- 71.Thacher S. M., Rice R. H. (1985) Cell 40, 685–695 [DOI] [PubMed] [Google Scholar]
- 72.Phillips M. A., Stewart B. E., Qin Q., Chakravarty R., Floyd E. E., Jetten A. M., Rice R. H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9333–9337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chakravarty R., Rong X. H., Rice R. H. (1990) Biochem. J. 271, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rice R. H., Green H. (1977) Cell 11, 417–422 [DOI] [PubMed] [Google Scholar]
- 75.Robinson N. A., Lapic S., Welter J. F., Eckert R. L. (1997) J. Biol. Chem. 272, 12035–12046 [DOI] [PubMed] [Google Scholar]
- 76.Pal R., Cristan E. A., Schnittker K., Narayan M. (2010) Biochem. Biophys. Res. Commun. 392, 567–571 [DOI] [PubMed] [Google Scholar]
- 77.Nickel W. (2003) Eur. J. Biochem. 270, 2109–2119 [DOI] [PubMed] [Google Scholar]
- 78.Nickel W., Rabouille C. (2009) Nat. Rev. Mol. Cell Biol. 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 79.Planey S. L., Keay S. K., Zhang C. O., Zacharias D. A. (2009) Mol. Biol. Cell 20, 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nickel W., Seedorf M. (2008) Annu. Rev. Cell Dev. Biol. 24, 287–308 [DOI] [PubMed] [Google Scholar]
- 81.Polakowska R. R., Eickbush T., Falciano V., Razvi F., Goldsmith L. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4476–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boutin J. A. (1997) Cell Signal. 9, 15–35 [DOI] [PubMed] [Google Scholar]
- 83.Boutin J. A., Ferry G., Ernould A. P., Maes P., Remond G., Vincent M. (1993) Eur. J. Biochem. 214, 853–867 [DOI] [PubMed] [Google Scholar]
- 84.Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 85.Younger J. M., Ren H. Y., Chen L., Fan C. Y., Fields A., Patterson C., Cyr D. M. (2004) J. Cell Biol. 167, 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Markossian K. A., Kurganov B. I. (2004) Biochemistry 69, 971–984 [DOI] [PubMed] [Google Scholar]
- 87.Candi E., Melino G., Lahm A., Ceci R., Rossi A., Kim I. G., Ciani B., Steinert P. M. (1998) J. Biol. Chem. 273, 13693–13702 [DOI] [PubMed] [Google Scholar]
- 88.Choate K. A., Williams M. L., Khavari P. A. (1998) J. Invest. Dermatol. 110, 8–12 [DOI] [PubMed] [Google Scholar]
- 89.Ahvazi B., Steinert P. M. (2003) Exp. Mol. Med. 35, 228–242 [DOI] [PubMed] [Google Scholar]
- 90.Ahvazi B., Boeshans K. M., Idler W., Baxa U., Steinert P. M. (2003) J. Biol. Chem. 278, 23834–23841 [DOI] [PubMed] [Google Scholar]
- 91.Breckenridge D. G., Germain M., Mathai J. P., Nguyen M., Shore G. C. (2003) Oncogene 22, 8608–8618 [DOI] [PubMed] [Google Scholar]