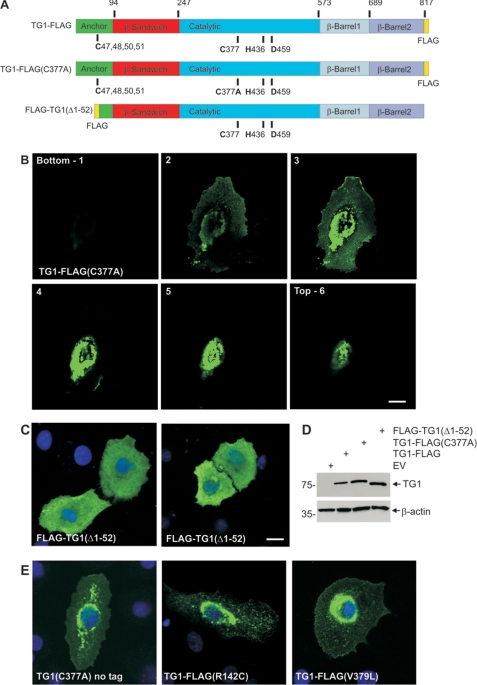
FIGURE 2.
Subcellular location of TG1 mutants. A, structure of TG1 showing the anchor, β-sandwich, catalytic, and β-barrel 1 and β-barrel 2 domains and the three residues that comprise the catalytic triad (Cys377, His435, and Asp459) and the four (C47,48,50,51) of the five cysteines that form the site that is lipid modified to add the membrane anchor. The position of the FLAG epitope tag is indicated. The numbers at the top indicate amino acids residue. The site of the mutation in TG1-FLAG(C377A) is indicated. FLAG-TG1(Δ1–52) is a truncation mutant. B, subcellular distribution of TG1-FLAG(C377A). Keratinocytes on coverslips were transfected with 2 μg of pcDNA3-TG1-FLAG(C377A) and after 48 h fixed and stained with anti-FLAG (green). The z-stack of images was generated with the bottom picture being at the coverslip/cell junction. C, cells were transfected with 2 μg of pcDNA3-FLAG-TG1(Δ1–52) and after 48 h fixed and stained with anti-FLAG. Nuclei were visualized using Hoescht stain. The bar = 10 μm in all panels. D, to monitor TG1-FLAG, TG1-FLAG(C377A) and FLAG-TG1(Δ1–52) expression cells were transfected with 10 μg of the indicated plasmid and after 48 h extracts were prepared for anti-FLAG immunoblot. E, cells were transfected with plasmids encoding TG1(C377A)(no tag), TG1-FLAG(R142C), and TG1-FLAG(V379L) and stained at 24 h with anti-TG1 (left panel) or anti-FLAG (middle and right panels). Nuclei were visualized using Hoescht stain. All images are 1-μm confocal optical sections.