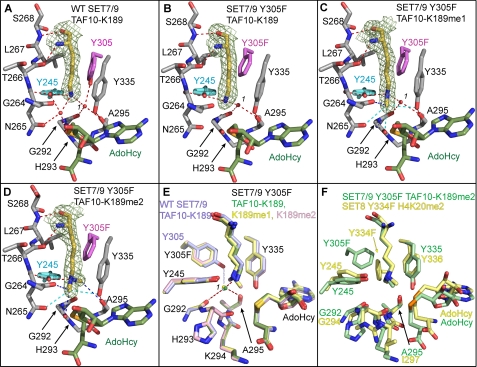
FIGURE 2.
Crystal structures of WT SET7/9 and the Y305F mutant in complex with AdoHcy and unmodified and methylated TAF10 peptides. A, active site of WT SET7/9 bound to AdoHcy and TAF10-K189 and the active site of SET7/9 Y305F bound to AdoHcy and TAF10-K189 (B), TAF10-K189me1 (C), and TAF10-K189me2 peptides (D). A–D, the K189 residue in the TAF10 peptide is depicted with yellow carbon atoms with the corresponding Fo – Fc simulated-annealing omit maps (contoured at 2.5 σ) shown. AdoHcy is colored with green carbon atoms, and SET7/9 carbon atoms are colored gray, with the exception of Tyr-305/Y305F (magenta) and Tyr-245 (cyan). Gly-264 to Ser-268 are shown, and the main chain atoms of Gly-292 to Ala-295 are depicted for clarity. Red dashed lines indicate conventional hydrogen bonds, and blue dashed lines indicate CH–O hydrogen bonds. Cyan dashed lines indicate weak CH–O hydrogen bonds between 3.5 and 3.7 Å in length. The water molecule in the solvent binding pocket is numbered 1. E, overlay of the active sites of the WT enzyme (light blue carbon atoms) and the Y305F mutant (green, yellow, and pink carbon atoms correspond to K189, K189me1, and K189me2, respectively). Red dashed lines indicate coordination of the waters in the structures of the Y305F mutant, and the light blue dashed line indicates hydrogen bonding that only occurs in the WT enzyme. F, superimposition of the active sites of the SET7/9 Y305F·AdoHcy·TAF10-K189me2 and SET8 Y334F·AdoHcy·H4K20me2 (Protein Data Bank code 3F9X) complexes shown with green and yellow carbon atoms, respectively.