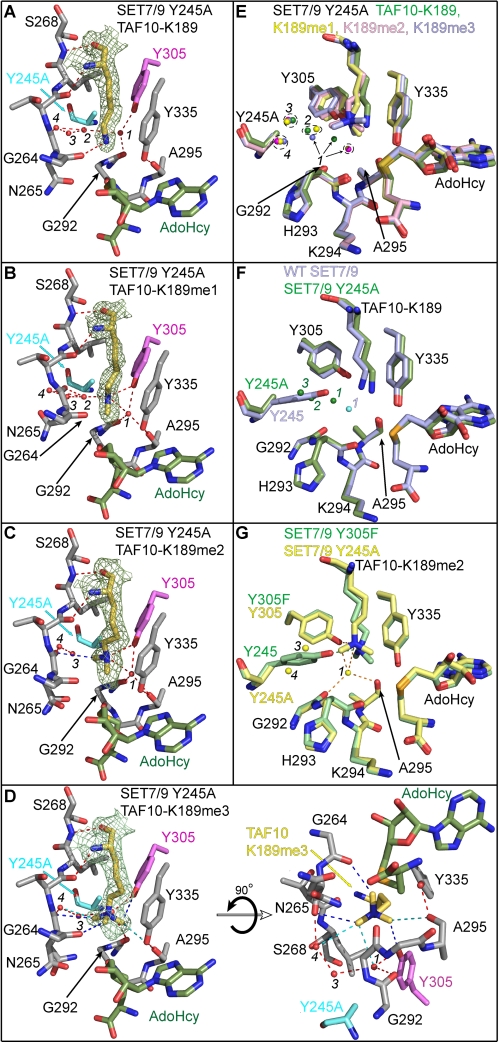
FIGURE 3.
Crystal structures of the SET7/9 Y245A mutant in complex with AdoHcy and unmodified and methylated TAF10 peptides. Active site of SET7/9 Y245A bound to AdoHcy and TAF10-K189 (A), TAF10-K189me1 (B), TAF10-K189me2 (C), and TAF10-K189me3 peptides (D). A–D, Fo – Fc simulated-annealing omit maps (contoured at 2.5 σ) for the unmodified and methylated K189 side chains are illustrated. The residues and hydrogen bonds in each complex are colored as in Fig. 2. The water molecules in the lysine binding channels of the Y245A complexes are numbered 1–4, as described in the text. E, superimposition of the active sites of the Y245A complexes bound to the four methylated states of TAF10-K189 (unmodified, mono-, di-, and trimethylated shown in green, yellow, pink, and blue, respectively). F, overlay of WT SET7/9 (blue carbons) and SET7/9 Y245A (green carbons) in complex with TAF10-K189. Waters corresponding to the WT and Y245A structures are colored cyan and green, respectively. G, alignment of the active sites of the Y245A (yellow carbons) and Y305F (green carbons) mutants in complex with the TAF10-K189me2 peptide. Hydrogen bonds from the Y305F structure are shown as green dashed lines, and waters and hydrogen bonds in the Y245A structure are shown in yellow and orange, respectively.