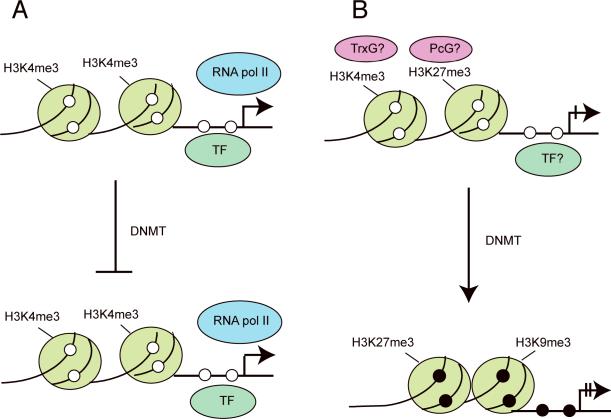
Figure 1. Targeted methylation or “passenger methylation” events.
A. Many genes are resistant to promoter methylation in cancer. Among the causes for this resistance can be association of a gene promoter with specific transcription factors or transcription factor complexes or presence of the activating histone mark H3K4me3, which interferes with DNA methylation.
B. Genes are targeted for methylation by a specific chromatin environment, for example lack of transcription and presence of the repressive Polycomb complex and the histone mark H3K27me3. These genes may initially be associated with `bivalent' chromatin characterized by both activating marks (H3K4me3 catalyzed by the Trithorax complex, TrxG) and repressive marks (H3K27me3 catalyzed by the Polycomb complex, PcG). In cancer tissue, these genes undergo DNA methylation and may be associated with other repressive marks (H3K9me3 and/or H3K27me3). The ball-shaped symbols represent nucleosome core particles over which the DNA is wrapped. Histone tail modifications are indicated and the open and closed circles represent unmethylated and methylated CpG sites, respectively.