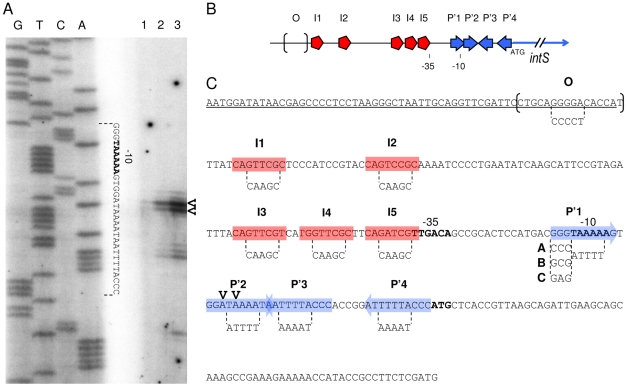
Figure 2. Primer extension analysis and mutagenesis strategy of the intS promoter.
A. The labeled primer was annealed to RNA extracted from MC4100 (lane 1), LCB1024 (ΔintS, lane 2) and LCB1019 (ΔKplE1 prophage) harboring pattL-gfp plasmid (lane 3) strains grown aerobically and extended with reverse transcriptase. Lanes G, T, C and A are a sequencing ladder of the attL DNA region. A complementary sequence is indicated between dotted lines. B. Schematic representation of IntS and TorI binding sites on attL that overlap the intS promoter region (I, TorI; P', IntS arm-type and O, IntS core-type). The −35 and −10 boxes and ATG of intS are indicated. C. intS promoter sequence. This sequence corresponds to the attL region cloned as a reference for PintS promoter studies (pattL-gfp, positions −223 to +64 relative to the ATG). The bold letters show the putative −35 and −10 boxes and ATG (2464.565 kb on the MG1655 E. coli chromosome) of the intS gene. Protein binding sites' sequences are indicated (red boxes, TorI; blue arrows, IntS arm-type; and brackets, IntS core-type) as well as the two initiation transcription sites (V). The mutagenesis of the protein binding sites was performed by overlap extension PCR to generate mutations (*). Substitutions are indicated between dotted lines. Contrary to the P'1* mutation that affects the consensus of IntS arm-type and the −10 box, P'1-(A, B or C) constructions do not affect the −10 box. The 3′ end of the argW tRNA gene sequence is underlined (13 nucleotides of argW are missing at the 5′ end).