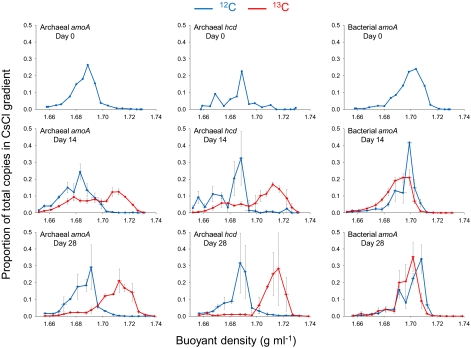
Fig. 3.
Buoyant density distribution of archaeal and bacterial genomic DNA extracted from soil after incubation with a headspace concentration of 5% (vol/vol) 12C- or 13C-CO2. Seventeen fractions from each CsCl spin were analyzed and represented a density range from approximatel 1.66 to 1.74 g mL−1. The abundances of archaeal amoA, archaeal hcd, and bacterial amoA genes in each fraction were determined by quantitative PCR, and values converted to the proportion of total gene abundance throughout the gradient. Vertical horizontal bars represent the SE of proportional abundance from triplicate CsCl spins (representing triplicate microcosms). Horizontal error bars represent the SE of the buoyant density of fractions collected from the same point in replicate CsCl tubes. Large vertical error bars observed in adjacent samples in some density gradients indicate that the largest gene abundance was detected in each of the adjacent fractions in replicate samples. Gene distribution profiles of day 0 samples (before incubation) are mean values derived from two individual CsCl gradients only.