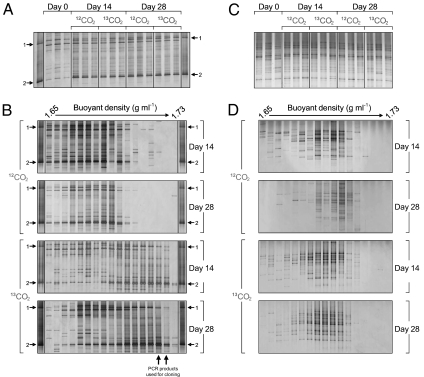
Fig. 4.
DGGE analysis of amoA gene PCR products from archaeal (A and B) and bacterial (C and D) amoA-defined communities derived from soil microcosms incubated for 14 or 28 d with a headspace concentration of 5% (vol/vol) 12C- or 13C-CO2. DGGE analysis was performed on amoA genes amplified from total genomic DNA from triplicate microcosms (A and C), with each lane representing an individual microcosm. DGGE analysis was also performed on 17 fractions recovered from individual CsCl density gradients separating genomic DNA of different buoyant densities from individual microcosms incubated for 14 or 28 d for both archaeal (B) and bacterial (D) amoA-defined communities. A marker lane containing PCR products from two group 1.1a clones (1 and 2, accession numbers FJ971889 and FJ971892, respectively) was run alongside all archaeal amoA DGGE profiles and represents sequences from organisms previously identified as growing under these incubation conditions. Two arrows indicate locations of archaeal amoA PCR products from two fractions that were pooled and used to construct clone libraries (Fig. S2).