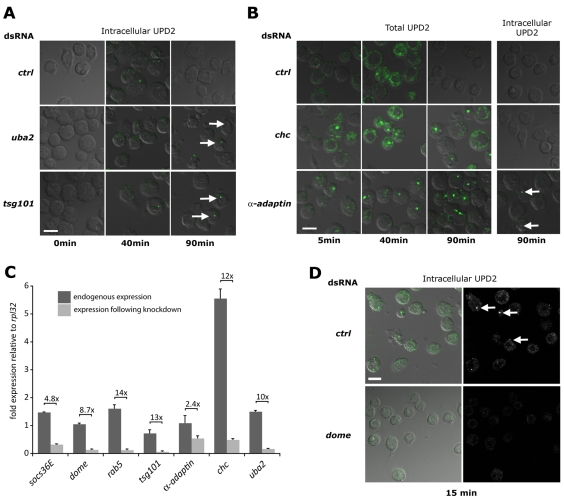
Fig. 3.
Endocytic gene knockdown and trapping of Upd2–Dome. Kc167 cells treated with the indicated dsRNAs before stimulation with Upd2-GFP (green) and incubation for the indicated times. Cells were either fixed directly (Total UPD2) or acid washed to remove external ligand (Intracellular UPD2). Cells are visualised by transmitted DIC illumination (grey). (A) Intracellular Upd2-GFP is clearly detectable within cells treated with control dsRNA following a 40 minute chase but cleared by 90 minutes. By contrast, cells treated with dsRNA targeting uba2 or tsg101 retain Upd2-GFP-labelled vesicles at 90 minutes (arrows). (B) Cells stained to visualise both cell surface and internalised Upd2-GFP are still associated with ligand following a 40 minute chase. Whereas wild-type cells treated with control dsRNA have cleared Upd2 after 90 minutes, cells treated with dsRNA targeting chc and α-adaptin retain Upd2-GFP (column 3). However, acid washing to remove extracellular Upd2-GFP reveals that no ligand is present within chc-knockdown cells, and only a few vesicles are present following knockdown of α-adaptin (arrows in column 4). (C) Quantitative-PCR of the indicated genes expressed in Kc167 cells showing fold expression relative to the ribosomal protein L32 gene (rpl32). Endogenous levels (dark bars) and expression following knockdown with dsRNA (light bars) are shown, as is the level of knockdown. All knockdowns are significant (P<0.001), except that of α-adaptin. (D) Intracellular Upd2-GFP is detected in cells treated with control dsRNA (arrows) but not in cells in which the domeless (dome) receptor is knocked down. Scale bars: 10 μm.