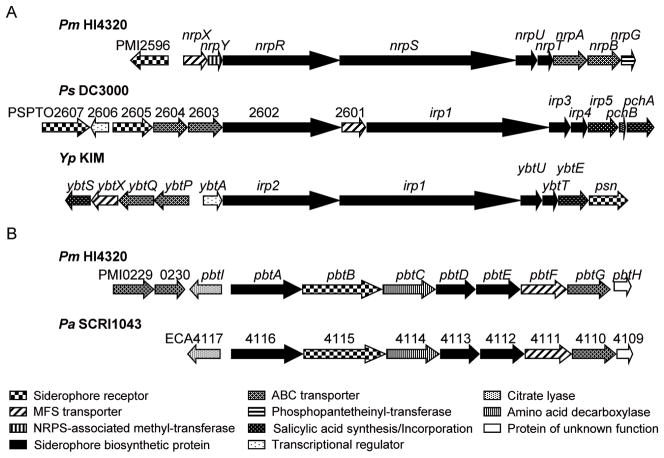
Fig. 4. The yersiniabactin-related siderophore and proteobactin of P. mirabilis are similar to siderophore biosynthesis and transport gene clusters in other Enterobacteriaceae as shown by gene sequence alignments.
(A) P. mirabilis yersiniabactin-related siderophore is homologous to that of P. syringae DC3000 and Y. pestis KIM. The yersiniabactin-related siderophore of P. mirabilis lacks the genes involved in salicyclic acid biosynthesis and incorporation, ybtS and ybtE of Y. pestis and irp5, pchB, and pchA of P. syringae. The nrp locus of P. mirabilis also lacks a transcriptional regulator but encodes a NRPS-associated methyl-transferase and a phosphopantetheinyl-transferase which are not located within the yersiniabactin operons of P. syringae and Y. pestis. (B) The newly identified NIS system of P. mirabilis, proteobactin, is similar to identical gene clusters found in P. carotovora subsp. atroseptica (shown) and P. carotovora subsp. carotovora.