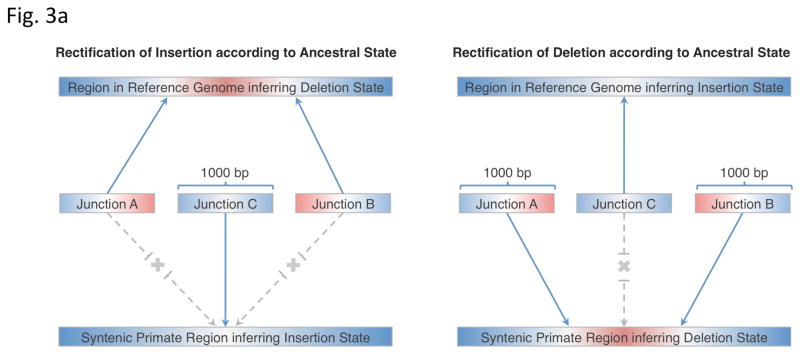
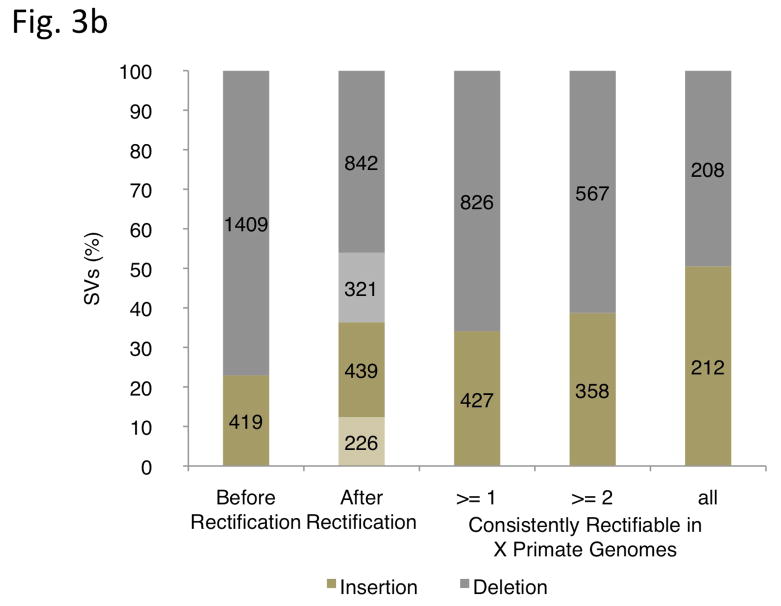
Figure 3.
Ancestral state classification. (a) Junction sequences are aligned onto syntenic regions of a non-human primate genome to infer SV ancestral states. For rectifying an SV insertion event (from deletion) according to ancestral state (left), sequences A and B represent the junction sequences of the reference SV allele, where as sequence C represents the junction sequence of the non-reference SV allele. For rectifying an SV deletion event (from deletion) according to ancestral state(right), sequence C represents the junction sequence of the reference SV allele and sequences A and B represent the junction sequences of the non-reference SV allele. Solid lines with arrows indicate successful alignments and dashed lines with crosses indicate no proper alignment. (b) Results of classifying SVs as insertions or deletions according to ancestral state. An SV event is defined as ‘rectifiable’ (indicated by darker color) if unambiguous high-quality alignments to putative ancestral regions could be constructed for the loci in any primate genomes (regardless of whether the classification is changed according to the ancestral state), and as ‘unrectifiable’ (represented by lighter color) if not.