Figure 6. NTPase activity of ABCE1 is required for its function in ribosomal recycling.
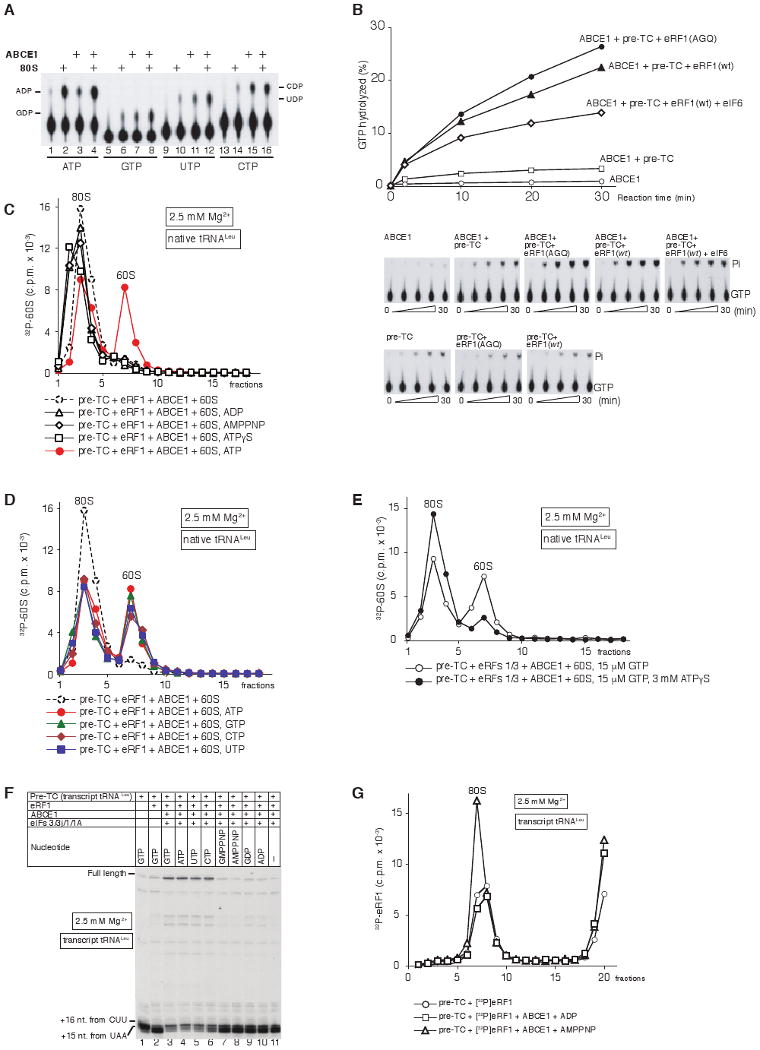
(A) Thin-layer chromatography analysis of ABCE1's NTPase activity in the presence/absence of 80S ribosomes. The positions of [α32P]-NDPs are indicated. (B) Time courses of GTP hydrolysis by ABCE1 in the presence/absence of pre-TC, pre-TC/eRF1(AGQ), pre-TC/eRF1(wt), or pre-TC/eRF1(wt)/eIF6, as indicated. 15 μl reaction mixtures containing 0.5 pmol ABCE1, 0.33 μM [γ-32P]GTP and combinations of 0.5 pmol pre-TC, 10 pmol eRF1(AGQ), 10 pmol eRF1(wt) and 10 pmol eIF6, were incubated at 37°C. Aliquots were removed after 2-30 minutes. GTP hydrolysis in the presence of both ABCE1 and pre-TCs (upper panels) was corrected to take into account the intrinsic GTPase activity of pre-TCs (lower panels). (C-E) Dissociation by ABCE1 of post-TCs, obtained by incubation of pre-TCs assembled on MVHL-STOP mRNA using native Leu-tRNALeu and [32P]60S subunits with (C, D) eRF1, or (E) eRF1/eRF3, depending on the presence/absence of nucleotides as indicated, assayed by SDG centrifugation. (F) Toe-print analysis of ribosomal complexes obtained by incubating pre-TCs assembled on MVHL-STOP mRNA using transcript tRNALeu with eRF1, ABCE1, eIFs and nucleotides, as indicated. The positions of full-length cDNA and of toe-prints corresponding to ribosomal complexes are indicated. (G) Association of [32P]eRF1 with post-TCs, assembled on MVHL-STOP mRNA with transcript Leu-tRNALeu, depending on the presence/absence of ABCE1 and nucleotides, assayed by SDG centrifugation. Upper fractions were omitted for clarity in panels C-E, G.
