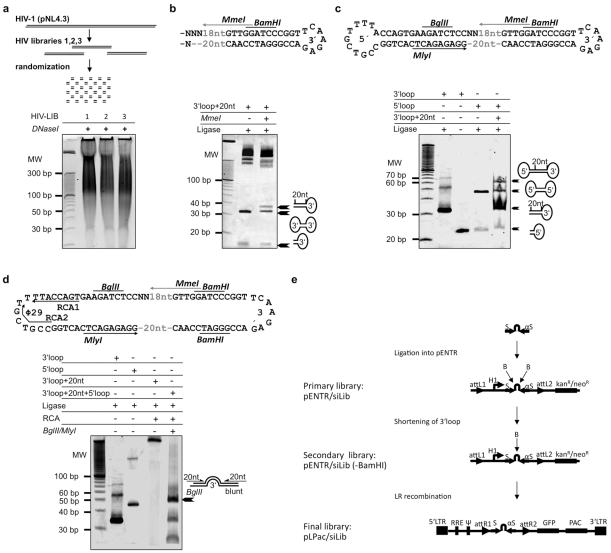
Figure 1. Construction of the shRNA-library.
a) The 3 HIV segments of pNL4.3 were fragmented using DNaseI and blunt-ended. b) Fragments of 150–300 bp were eluted and ligated to the 3′ loop. To limit the size of the HIV-1 inverted repeats, a recognition site for MmeI, which cleaves exactly 20nt from its recognition site and leaves 2 nt 5′ overhangs, was engineered into the 3′ loop. c) Ligation of the 5′ loop to the MmeI-digested fragments generated a quasi-circular single-stranded structure. d) Rolling circle amplification (RCA) reactions using Φ 29 DNA polymerase and the primers RCA1 and RCA2 were performed to amplify the single-stranded circular DNA and to generate the complementary strand yielding a DNA concatemere of palindromic, inverted repeats encoding siRNA molecules. Digestion with BglII and MlyI liberated shRNA sequences which were inserted into the expression vector pENTR/siLib. d) The shRNA sequences were cloned into the linearized pENTR/Lib generating the primary library which was religated after BamHI digestion yielding the secondary library. The final lentiviral shRNA-library was generated by LR recombination of the secondary library into pL/EGP/siLib.