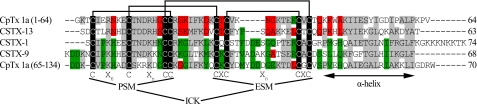
FIGURE 3.
CpTx 1 sequence homologues. Two modules (domains) of CpTx 1 corresponding to amino acid residues 1–64 and 65–134 are aligned with sequences of toxins from C. salei CSTX-1, -9, and -13. Polypeptide chain lengths are given in the right column. Note that CSTX-13 is a two-chain molecule, with no covalent bond between Gln-34 and Ala-35. Similar residues are shaded gray. Residues more common to the N-terminal domain of CpTx 1 and CSTX-13 are highlighted red; those more common to the C-terminal domain of CpTx 1 and toxins CSTX-1 and 9 are highlighted green. Invariant cysteine residues are shaded black, and the disulfide bonds assigned for CSTX toxins are drawn above. The cysteine motifs the principal structural motif (PSM) and extra structural motif (ESM), each contributing to the ICK structural fold, are identified below. The putative C-terminal α-helical region is also indicated below.