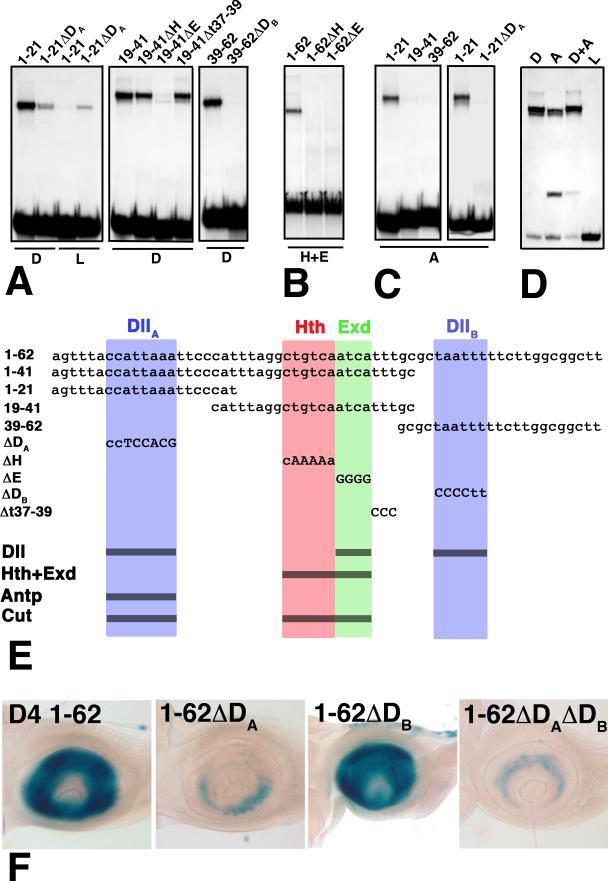
Figure 4.
Gel-shift assays of mutant and wild-type derivatives of D4. The sequences of full length D4, four fragments, and five clustered site mutations are shown in (E). Abbreviations as in Fig. 3. (A) Dll binds three sites in D4, one in each of the subfragments 1-21, 19-41, and 39-62. Binding to these fragments is almost completely eliminated by the ΔDA, ΔE, and ΔDB mutants, respectively. (B) Cooperative binding of Hth and Exd is eliminated in both the ΔH and ΔE mutants, indicating that Hth and Exd bind adjacent consensus sites. (C) Antp binds only the 1-21 fragment, and this binding is lost in the ΔDA mutant. Lysate (L) control lanes were blank (not shown) for all but the 1-21ΔDA probe. (D) Purified Dll (D) and Antp (A) compete for binding to the 1-21 probe. The faster migrating band in the Antp lanes is likely due to the binding of a breakdown product generated during purification. (E) Summary of the DNA sequences tested in (A-D). (F) Effects of the ΔDA and ΔDB mutations on antennal expression in vivo.