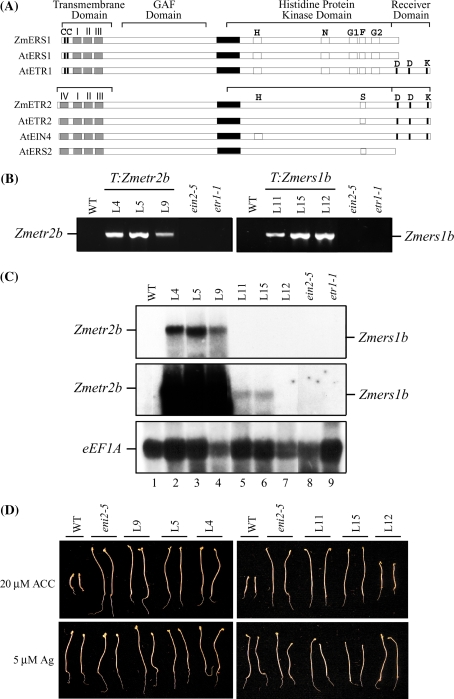
Fig. 1.
Zmetr2b and Zmers1b function as dominant negative mutants in Arabidopsis. (A) Comparison of ZmERS1 with Arabidopsis subfamily I receptors, i.e., AtETR1 and AtERS1, and comparison of ZmETR2 with Arabidopsis subfamily II receptors, i.e., AtETR2, AtEIN4, and AtERS2. The N-terminal, hydrophobic, transmembrane domains are indicated by gray boxes. Cys-4 and Cys-6, are indicated by the Cs at the left end of the proteins. The five consensus motifs (H, N, G1, F, and G2) within the histidine protein kinase domain (Hua et al. 1998) are indicated, and the aspartate and lysine residues conserved in the receiver domain of ETR1 are indicated. The serine-rich domain (S) is also indicated. The proposed coiled-coil region is indicated by the black box. (B) PCR amplification of the Zmetr2b and Zmers1b transgenes from three lines containing Zmetr2b (i.e., L4, L5, L9) and three lines containing Zmers1b (i.e., L11, L15, L12) confirming the presence of the transgene in the transformants. Wild-type (WT), ein2-5, and etr1-1 plants were included as negative controls. (C) Northern analysis of seedlings of the same Zmetr2b or Zmers1b lines germinated in the dark for 10 days. The level of Zmetr2b and Zmers1b expression was measured using a mixture of Zmetr2b and Zmers1b probes after a 24 h (top panel) or 2 week (middle panel) exposure of the membrane to film. Expression of the translation elongation factor 1A (eEF1A) mRNA was determined as an RNA loading control from the same membrane after it had been stripped (bottom panel). (D) Seeds from the same lines were germinated in the dark for 5 days on media containing either 20 μM ACC or 5 μM AgNO3 to assay for their triple response. Two representative seedlings are shown for each line. Quantitative measurements for hypocotyl and root lengths with standard deviations are shown in Table 1