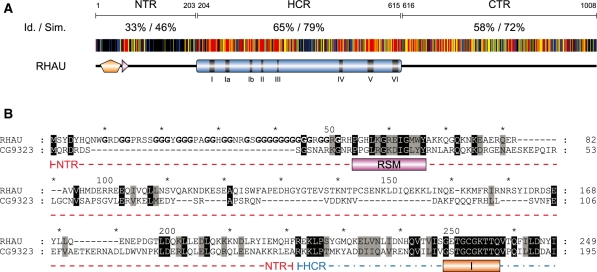
Figure 8.
Amino acid conservation of the N-terminal region of RHAU. (A) Schematic representation of amino acid conservation of human RHAU throughout evolution. The human RHAU sequence was aligned with eight RHAU orthologues (the sequences shown in Figure 6) by MAFFT (version 6) (35). Each residue of RHAU is represented with a colour code that indicates its level of conservation amongst the eight orthologous sequences. Similarity is shown in red for 100%, yellow for 99–80% and blue for 79–60%. Similarity analysis was made by GeneDoc (version 2.7) using the BLOSUM62 scoring matrix. Average values of identity (Id) and similarity (Sim) for N-terminal (NTR), helicase core (HCR) and C-terminal (CTR) regions are indicated. (B) Sequence alignment of the N-terminal region of RHAU with its Drosophila ortholog CG9323. Amino acids that are identical or similar between the two sequences are shaded in black and grey, respectively. The RSM domain as well as helicase motif I are indicated below the sequences. Gly residues of the Gly-rich domain (amino acid 10–51) of RHAU are bolded. N-terminal region and a part of the helicase core domain are delineated with a coloured dashed line in red and blue, respectively. For complete alignment between RHAU and CG9323, see Supplementary Figure S5.