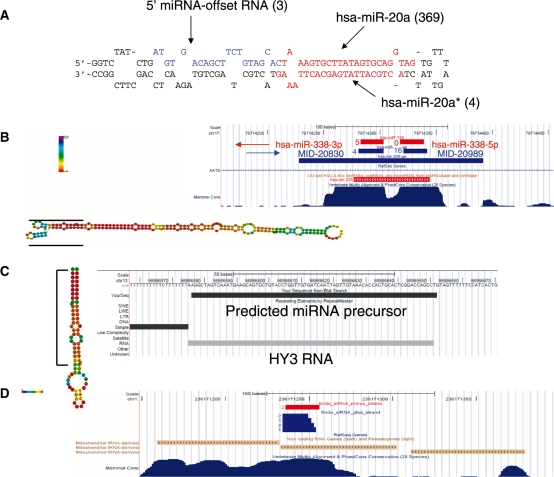
Figure 3.
Novel types of small RNA identified in deep sequencing of tumors. (A) 5′ miRNA-offset RNA from hsa-mir-20a. In parenthesis are numbers of sequenced reads. (B) Antisense miRNA and miRNA star of hsa-miR-338. Red segments denote hsa-miR-338 precursor and mature miRNAs (long segments are precursors); blue segments denote the antisense miRNA-338 precursor and mature miRNAs. Numbers beside the short segments (mature miRNA) denote number of sequenced reads. The predicted folded hairpin of the antisense miRNA precursor is shown on the left with the positions of the mature miRNAs marked by black lines. Nucleotides colored in red have a high probability to occupy the depicted fold, green colored nucleotides have a low probability to be in the given fold. (C) miRNA sized Small RNA derived from Y-RNA. The entire Y RNA is folded as a hairpin, similar to miRNA precursor (left). The sequenced small RNA is derived from the 5′ arm of the hairpin. (D) Putative endogenous siRNA derived from mitochondrial-derived pseudogene. Segments in blue derive from plus strand and start in the transcription start site of the tRNA pseudogene. Segments in red derive from the minus strand. Numbers beside the segments denote number of sequenced reads. This figure is based on the UCSC genome browser (53).