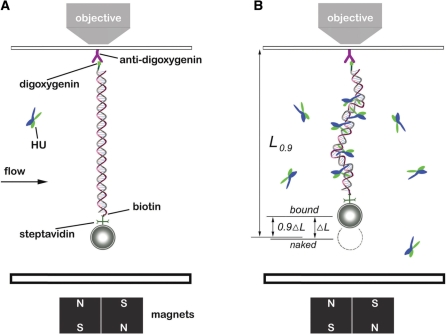
Figure 1.
Experimental configuration. (A) Schematic of magnetic tweezers setup. DNAs end-labeled with biotin and digoxygenin were attached to paramagnetic microbeads through biotin-streptavidin cross-linkers. Then the bead-DNA constructs were adhered to a piece glass slide on a flow cell by digoxygenin-antibody linkage. The images were magnified and tracked by a piezo controlled 100× objective, and captured by a CCD camera. The bead was manipulated by motor-driven permanent magnets near the flow cell. The solution inside the flow cell could be changed, allowing HU–DNA association and dissociation to be controlled. (B) DNA after HU binding. The extension of the HU–DNA complex (Lbound) was shorter than the original naked DNA length (Lnaked) due to bending of DNA by the proteins. The difference of Lnaked and Lbound was denoted by ΔL and taken as an indicator of amount of protein bound. During protein dissociation experiments, the time t0.9 for the extension to return 90% of the way back to Lnaked (to the level L0.9), was used to measure the complex dissociation lifetimes.