Abstract
The role of curli, amyloid extracellular fibers, in the tolerance of Escherichia coli PHL628 to Hg(II) was examined. Our findings indicate that by sorbing Hg(II) extracellularly, curli protect the cells. To our knowledge, this is the first time a protective role of curli against toxic metals has been demonstrated.
Mercury is a toxic metal with no known biological function because its affinity to thiol compounds is so high that it disrupts protein structure and function (18). Bacteria are involved in the global environmental cycling of mercury. They both reduce Hg2+ to metallic Hg0, which is volatile and relatively inert, and methylate ionic Hg(II) in an enzymatic process that makes Hg more bioavailable and more toxic (1). Bacteria may cope with heavy metal stress by actively exporting metal cations from the cytoplasm (17). Additionally, the binding of heavy metals by extracellular polymeric substances has been demonstrated to be an effective way to increase the resistance to these toxic cations (5, 8, 11, 16, 19). However, other less understood mechanisms of tolerance are also likely to exist.
Curli are thin, aggregative, extracellular fibers produced by many enterobacteria (13) but which are also abundant in natural biofilms, particularly in oligotrophic environments (14, 15). The genes responsible for the production of curli in Escherichia coli are clustered in two divergent operons: the csgBA operon, which encodes the major structural subunit, CsgA, and the csgDEFG operon (7). The regulation of curli gene expression is complex and responsive to environmental cues, such as cell wall stress (20), but in general, curli genes are maximally expressed during stationary phase (2).
E. coli PHL628, a curli-deficient derivative (PHL628 csgA), and its plasmid-borne csgA complement [PHL628 csgA(pBBR1MCS3:csgA)] were used as the test organisms for the work described in this report. PHL628 is a K-12 MG1655 derivative with a single point mutation in the regulatory protein OmpR (21). This allele enhances curli gene expression and promotes biofilm formation (21). Strain and plasmid details are listed in Table 1, and primers used in this study are listed in Table 2. The ability of the csgA-deficient strain to form curli was restored by cloning the csgA gene in the vector pBBR1MCS3 (12), followed by transformation of the cells with pBBR1MCS3:csgA following the protocol described by Choi et al. (3). The presence or absence of curli was verified using an autoaggregation assay (10) (data not included).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description and relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| PHL628 | MG1655 malA-Kan ompR234 Kanr | 21 |
| PHL628 csgA | csgA deficient; Kanr Cmr | 20 |
| Plasmids | ||
| pBBR1MCS3 | Broad-host-range cloning vector; Tetr | 12 |
| pBBR1MCS3:csgA | pBBR1MCS3 with csgA gene insert; Tetr | This study |
| pmerRGFP | Hg uptake reporter vector; Ampr | 6 |
TABLE 2.
PCR primers used in this study
Cultures were grown in a glucose minimal medium (GMM) which contained, per liter, 7 g K2HPO4, 2 g KH2PO4, 1 g (NH4)2SO4, 200 mg MgSO4·7H20, 24 mg Ca(NO3)2·4H20, and 10 g glucose. The medium was buffered at pH 7.2 with HEPES. Analytical-grade HgCl2 was obtained from Mallinckrodt, and stock solutions were 2% acidified with trace metal-grade HNO3.
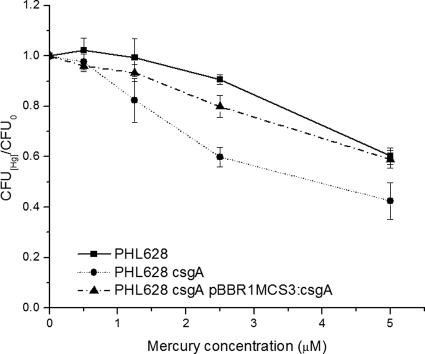
The role that curli play in the resistance to Hg(II) was tested by exposing stationary-phase cultures of PHL628, PHL628 csgA, and PHL628 csgA(pBBR1MCS3:csgA) to various amounts of Hg(II). After 3 h of incubation, dilutions of the cell suspensions were plated and CFU were counted. The decrease in CFU with increasing mercury concentration was more pronounced in the curli-deficient strain, and mercury resistance was restored by complementation of the deletion (Fig. 1). This result led us to conclude that curli provide moderate protection against mercury toxicity.
FIG. 1.
Viability curves. CFU of PHL628(pBBR1MCS3), PHL628 csgA, and PHL628 csgA(pBBR1MCS3:csgA) exposed to Hg(II) divided by the CFU of unexposed cultures plotted versus Hg concentration. Cultures were grown for 24 h at 30°C in GMM, spiked with Hg(II), and incubated for 3 additional hours before the dilutions were prepared and plated. Error bars represent standard deviations of the results for triplicate samples.
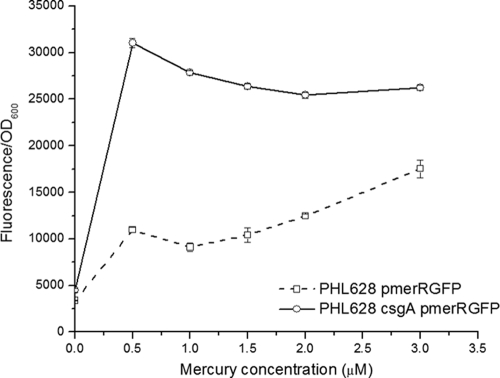
Next, we assessed the potential for curli to alter the uptake of Hg(II). Fluorescence may be used as a surrogate measurement for mercury uptake in cells that carry pmerRGFP (6) because fluorescence is produced only when Hg(II) enters the cytosol (4). Cultures of PHL628 and PHL628 csgA harboring pmerRGFP were grown to stationary phase at 30°C, and the bacteria were then exposed to various Hg(II) concentrations. Hg-induced fluorescence was normalized to cell concentration as measured by optical density at 600 nm (OD600) in order to account for the variability in cell numbers. We found that the curli-deficient strain had a higher normalized fluorescence (fluorescence/fluorescence at OD600) than the wild-type strain over the range of concentrations tested (Fig. 2).
FIG. 2.
Hg uptake. PHL628 and PHL628 csgA cultures harboring pmerRGFP grown for 24 h in GMM at 30°C were harvested and exposed to total Hg concentrations ranging from 0 to 3 μM. The green fluorescence and OD600 values of the cultures were recorded for the following 4 h. For each condition, the fluorescence was divided by the OD600, and the resulting values from three independent experiments were averaged and plotted versus time. Error bars represent standard deviations of the results for triplicate samples. Results at 4 h after Hg introduction are shown.
Our results support the idea that curli protect the bacteria from exposure to Hg(II) by reducing uptake. The simplest explanation for this result is that curli sequester the metal in the sheath they form on the outside of the cell, consequently preventing or delaying mercury from entering the cell.
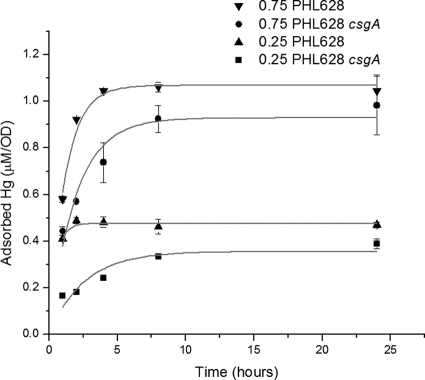
To provide additional evidence in support of this hypothesis and study the role of curli in Hg adsorption, we exposed stationary-phase cells of PHL628 and PHL628 csgA to a total Hg concentration of 0.25 or 0.75 μM and determined the amount of Hg that sorbed to the cells at 1, 2, 4, 8, and 24 h by calculating the difference with the following formula: total Hg − soluble Hg = sorbed Hg (Fig. 3). Soluble Hg concentrations were determined using a Bacharach Coleman model 50B analyzer system (Bacharach, Inc., Pittsburgh, PA) following a cold vapor atomic absorption method (9). The data for adsorbed Hg per cell were used to fit an adsorption model with a limited number of binding sites. Quantification of the difference in adsorption kinetics relative to curli was done by modeling the adsorption process as the following first-order differential equation, where binding is limited by the number of available binding sites: dC/dt = k(C* − C), with the solution C(t) − C*(1 − e−kt), where C is the Hg concentration adsorbed to cells divided by the OD600 measured at each time point (μM/OD), t is time (h), k is the binding rate constant (h−1), and C* is the maximum Hg concentration adsorbed to cells at equilibrium (μM/OD).
FIG. 3.
Kinetics of Hg adsorption to PHL628 strains at pH 7.2. Suspended cultures of PHL628 and PHL628 csgA were grown in GMM at 30°C to stationary phase. Aliquots of concentrated Hg solution were added to the cultures to yield total concentrations of 0.25 μM and 0.75 μM Hg. Aliquots were withdrawn at predetermined intervals, and soluble Hg concentrations were determined. Adsorbed Hg normalized by biomass was calculated as the difference between the total added and dissolved Hg concentrations, and after correcting for background values, it was divided by the OD600 and plotted versus time. The points and whisker bars indicate the average and the range of the results for duplicate samples; the gray lines represent the calculated fit.
The experimental adsorption data were fitted to the integrated rate equation using a Levenberg-Marquardt nonlinear fitting algorithm in OriginPro8 (Northampton, MA), where C* and k were used as the only fitting parameters. Quantitatively, our results indicate that curli speed up the kinetics of adsorption. Also, more mercury adsorbed to cells with curli at equilibrium (Table 3 and Fig. 3). At 0.75 μM Hg(II), the magnitude of the fitted rate constant was approximately 40% lower for the curli-deficient strain. At 0.25 μM Hg(II), the rate constant was approximately 80% lower.
TABLE 3.
Hg adsorption kinetics constants
| Strain | Total Hg exposure (μM) | k (h−1) | Range | C* (μM/OD) | Range |
|---|---|---|---|---|---|
| PHL628 | 0.25 | 2.00 | 0.30 | 0.48 | 0.01 |
| PHL628 csgA | 0.25 | 0.40 | 0.10 | 0.36 | 0.05 |
| PHL628 | 0.75 | 0.85 | 0.07 | 1.07 | 0.03 |
| PHL628 csgA | 0.75 | 0.52 | 0.09 | 0.93 | 0.06 |
In this work, we have shown that curli provide modest protection against mercury toxicity and appear to do so by sequestering the metal extracellularly, thereby reducing the amount that is bioavailable. This effect is likely a result of the increased curli-associated extracellular surface area available for metal adsorption and is based on the following observations: (i) curli-forming strains were more resistant to toxic mercury concentrations than the curli-deficient strain (Fig. 1), (ii) the uptake of Hg(II) increased in the absence of curli (Fig. 2), and (iii) bacteria without curli adsorb less Hg than curli-forming bacteria (Fig. 3). Curli will protect cells temporarily by slowing the diffusion of transient increases in bulk-solution Hg concentration to the cell wall and will protect cells on a long-term basis as long as there is an excess in the ratio of curli adsorption sites to Hg. Once these sites are saturated, the protective effect of curli will disappear. To our knowledge, this is the first time that curli have been proven to protect bacteria from metal toxicity. Although more work is required in order to understand the exact mechanism of Hg(II) tolerance, the protective effect afforded by curli may contribute to the widespread occurrence of these structures in enteric bacteria and environmental biofilms.
Acknowledgments
This research was supported in part by NSF grant EAR-0311767. Partial support for Gabriela Hidalgo was provided by the College of Engineering at Cornell University.
We thank M. Virta (University of Turku, Finland) for plasmid pmerRGFP. We are grateful to J. M. Moran-Mirabal for help with the adsorption model.
Footnotes
Published ahead of print on 20 August 2010.
REFERENCES
- 1.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 2.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, K., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 4.Golding, G. R., C. A. Kelly, R. Sparling, P. C. Loewen, and T. Barkay. 2007. Evaluation of mercury toxicity as a predictor of mercury bioavailability. Environ. Sci. Technol. 41:5685-5692. [DOI] [PubMed] [Google Scholar]
- 5.Gutnick, D. L., and H. Bach. 2000. Engineering bacterial biopolymers for the biosorption of heavy metals; new products and novel formulations. Appl. Microbiol. Biotechnol. 54:451-460. [DOI] [PubMed] [Google Scholar]
- 6.Hakkila, K. 2002. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 301:235-242. [DOI] [PubMed] [Google Scholar]
- 7.Hammar, M., A. Arnqvist, Z. Bian, A. Olsén, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 8.Harrison, J. J., R. J. Turner, and H. Ceri. 2005. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ. Microbiol. 7:981-994. [DOI] [PubMed] [Google Scholar]
- 9.Hatch, W. R., and W. L. Ott. 1968. Determination of submicrogram quantities of mercury by atomic absorption spectrophotometry. Anal. Chem. 40:2085-2087. [Google Scholar]
- 10.Hidalgo, G. 2010. Aspects of transition metal interaction with bacteria: imaging of extracellular pH in biofilms, Hg binding to curli, and differential gene expression in response to Hg. Ph.D. thesis, Cornell University, Ithaca, NY.
- 11.Kaplan, D., D. Christiaen, and S. Arad. 1987. Chelating properties of extracellular polysaccharides from Chlorella spp. Appl. Environ. Microbiol. 53:2953-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovach, M. E., P. H. Elzer, D. Steven Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 13.Landini, P., G. Jubelin, and C. Dorel-Flamant. 2006. The molecular genetics of bioadhesion and biofilm formation. In Biological adhesives, 1st ed. Springer, Heidelberg, Germany.
- 14.Larsen, P., J. L. Nielsen, M. S. Dueholm, R. Wetzel, D. Otzen, and P. H. Nielsen. 2007. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9:3077-3090. [DOI] [PubMed] [Google Scholar]
- 15.Liss, S. N., I. G. Droppo, D. T. Flannigan, and G. G. Leppard. 1996. Floc architecture in wastewater and natural riverine systems. Environ. Sci. Technol. 30:680-686. [Google Scholar]
- 16.McLean, R. J. C., D. Beauchemin, L. Clapham, and T. J. Beveridge. 1990. Metal-binding characteristics of the gamma-glutamyl capsular polymer of Bacillus licheniformis ATCC 9945. Appl. Environ. Microbiol. 56:3671-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 18.Nies, D. H. The elements: essential and toxic effects on microorganisms. In Elements and their compounds in the environment, 2nd ed. Wiley-VCH, Weinheim, Germany.
- 19.Teitzel, G. M., and M. R. Parsek. 2003. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 69:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toba Francis, F. A. 2008. Characterization of the physiological implications of defective lambdoid phage DLP12 in E. coli. Ph.D. thesis, Cornell University, Ithaca, NY.
- 21.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]