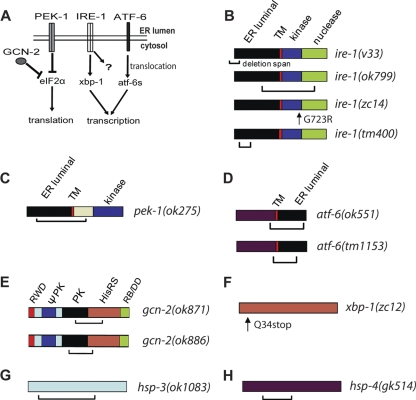
FIG. 3.
UPR pathway and mutants. (A) Schematic of UPR pathways. PEK-1, IRE-1, and ATF-6 are activated in the presence of unfolded proteins in the ER lumen. These pathways can promote adaptation to unfolded proteins via translational suppression or through a transcriptional response. GCN-2 functions along with activated PEK-1 to suppress translation. (B) ire-1 mutations. ire-1(v33) has an N-terminal 878-bp deletion resulting in a frameshift and stop and is a presumptive null mutation (48). ire-1(ok799) has a 2,093-bp deletion and 409-bp insertion and also should represent a null mutation (50). ire-1(zc14) has a missense mutation in a conserved residue in the kinase domain (8). ire-1(tm400) has a 600-bp deletion and 1-bp insertion that ends in an intron (see Wormbase.org and Materials and Methods). The mutant product is unclear. (C) pek-1(ok275) has a 2,073-bp deletion that produces a frameshift and stop codon (48). (D) Proteolysis of ATF-6 produces ATF-6s with only the maroon domain that is truncated by both mutations. ok551 has a 1,900-bp deletion (49); tm1153 has a 643-bp frameshift deletion (Wormbase.org). (E) gcn-2(ok871) has a 1,481-bp in-frame deletion starting and ending in exons (33); gcn-2(ok886) has a 1,179-bp in-frame deletion that starts and ends in exons (33). (F) xbp-1(zc12) has an early stop (8). (G) hsp-3(ok1083) has a 1,422-bp deletion that starts and ends in exons, causing frameshift (22). (H) hsp-4(gk514) has a 752-bp deletion that starts and ends in exons, causing frameshift (46). TM, transmembrane domain. The RWD domain was named after three major RWD-containing proteins: RING finger-containing proteins, WD-repeat-containing proteins, and yeast DEAD (DEXD)-like helicases. ΨPK, degenerate kinase domain; PK, kinase domain; HisRS, histidyl-tRNA synthetase; RB/DD, ribosome-binding and dimerization domain.