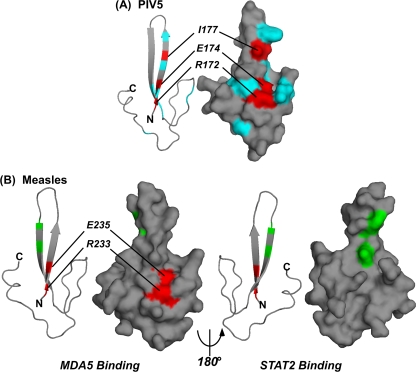
FIG. 6.
Structural models for paramyxovirus V protein CTD contact residues. (A) The PIV5 V protein CTD derived from the crystal structure (PDB accession no. 2B5L), colorized to represent the mutations described here. For simplicity, the zinc atoms are not shown, and their coordinating residues are not colored. Mutations that do not alter MDA5 interference are represented in cyan (R173, S176, W179, W189, N191, P192, I197, T198, and P212). Mutations that impair MDA5 interference are represented in red and labeled. (B) The measles virus V protein sequence was modeled on the simian virus 5 structure (PDB accession no. 2B5L) by use of the Swiss PDB viewer available at http://www.expasy.org/spdbv/ (15). Each pair of images illustrates a ribbon view on the left and a surface view on the right, with front and back faces indicated in a 180° rotation. Coloring corresponds to the residues identified as important for the measles virus V protein association with MDA5 (red) or those important for association with STAT2 (green) (F246R and D248F) (42).