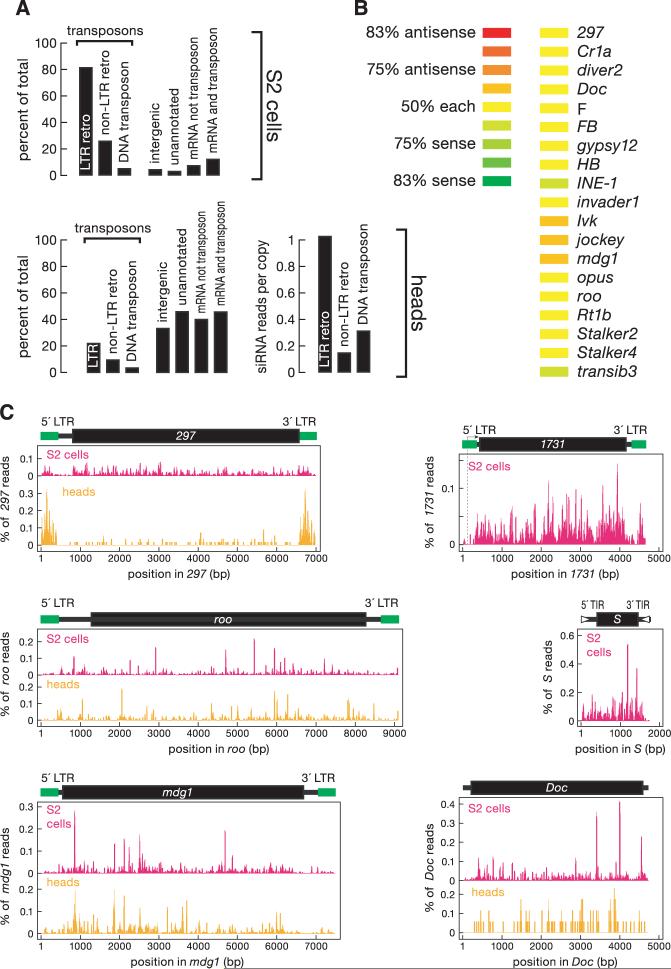
Fig. 2.
Endo-siRNAs correspond to transposons. (A) Distribution of annotations for the genomic matches of endo-siRNA sequences. Bars total more than 100% because some siRNAs match both LTR and non-LTR retrotransposons or match both mRNA and transposons. (B) Transposon-derived siRNAs with more than 50 21-nt reads mapped about equally to sense and antisense orientations. (C) Alignment of endo-siRNA sequences to Drosophila transposons. The abundance of each sequence is shown as a percentage of all transposon-matching siRNA sequences. LTR, long terminal repeat; TIR, terminal inverted repeat. Here and in subsequent figures, data from high-throughput pyrosequencing and sequencing-by-synthesis were pooled for wild-type heads.