Figure 4. Arabidopsis AtVMA21a and AtVMA21b demonstrate ER localization similar to the yeast V-ATPase assembly factor Vma21p.
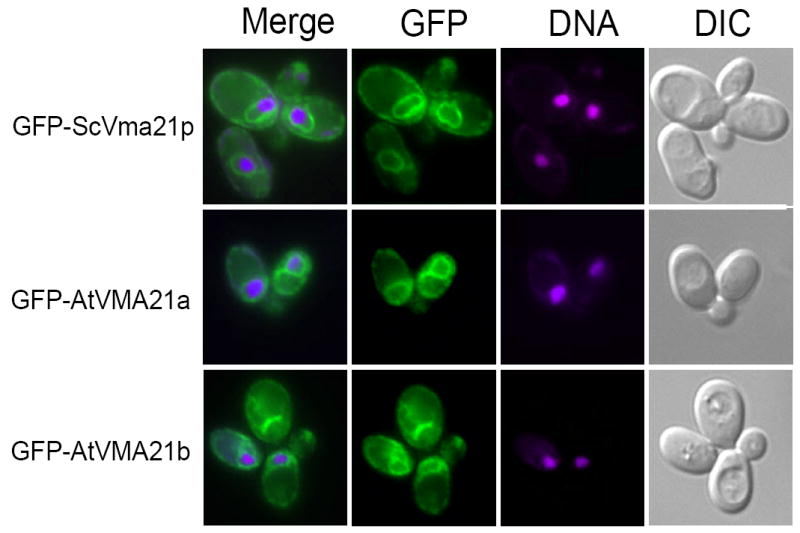
Cell imaging of vma21Δ yeast cells transformed with N-terminal GFP-tagged constructs of ScVma21p (pLG278), AtVMA21a (pLG269) or AtVMA21b (pEB05). Prior to cell imaging samples were fixed (3.7% formaldehyde for 10 minutes) and incubated with 5 mg/ml Hoechst 33342 DNA binding dye. The panels on the right show yeast cell morphology as viewed by differential interference contrast microscopy (DIC). The panels on the far left are a merge of the green GFP fluorescence (second set of panels from the left) with the magenta-stained nucleic acid fluorescence (third set of panels from the left).
