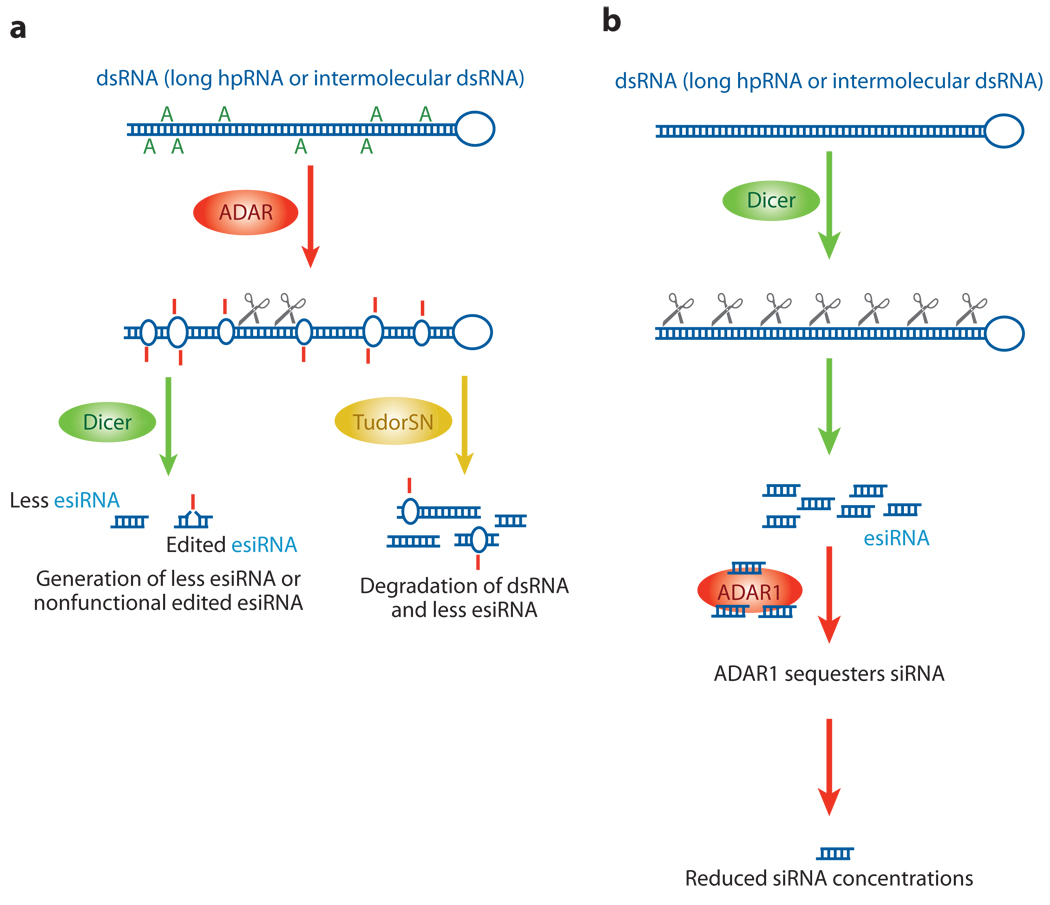
Figure 4.
Interaction between RNA editing and RNAi pathways. Two possible ways in which RNA editing and RNAi pathways may interact. (a) The introduction of many I·U base pairs and the alteration of the dsRNA structure by ADAR leads to the generation of fewer esiRNAs by Dicer, because dsRNAs that contain many I·U base pairs become resistant to Dicer cleavage. Alternatively, extensively edited dsRNAs with multiple I·U base pairs may be rapidly degraded by Tudor staphylococcal nuclease (Tudor-SN), resulting in the generation of fewer esiRNAs. (b) In addition, a fraction of already processed siRNAs might be sequestered by certain ADAR gene family members, reducing the effective siRNA concentration. For instance, cytoplasmic ADAR1p150 binds siRNA tightly. Gene silencing by siRNA is significantly more effective in the absence of ADAR1, indicating that ADAR1p150 is a cellular factor that limits siRNA potency in mammalian cells by decreasing the effective siRNA concentration and its incorporation into RNA-induced silencing complex.