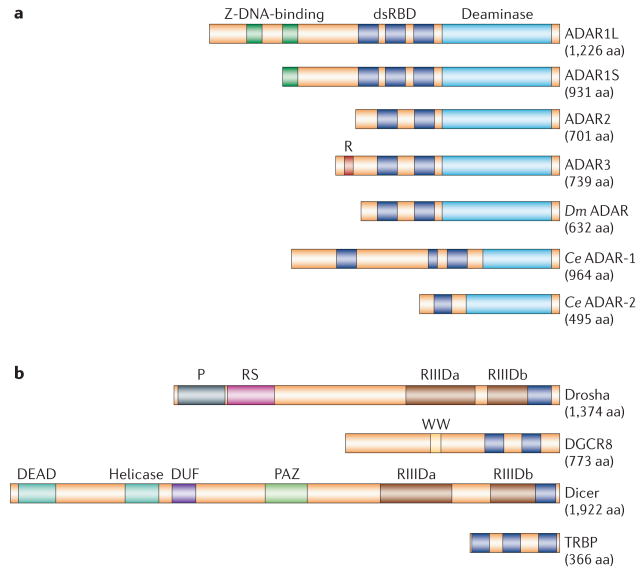
Figure 2. Types of dsRBD-containing protein: ADAR-family proteins and proteins that are required for miRNA biogenesis.
a | Three human ADAR (adenosine deaminase acting on RNA)-family members (ADAR1–3), Drosophila melanogaster (Dm) ADAR and two Caenorhabditis elegans (Ce) proteins, ADAR-1 and ADAR-2, share common functional domains: 2 or 3 repeats of the dsRBD and a catalytic deaminase domain. Certain structural features, such as Z-DNA-binding domains and the Arg-rich (R) domain, are unique to particular ADAR members. Binding of ADAR to double-stranded (ds)RNA substrates is mediated through dsRBDs38, whereas Z-DNA-binding domains might increase the affinity of ADAR1L specifically for short dsRNAs such as siRNAs22. Binding of the R domain to single-stranded RNAs has been reported, but its biological significance is currently unknown35. Two ADAR1 translation products, the isoforms ADAR1L and ADAR1S, result from transcription from different promoters followed by alternative splicing. This leads to translation initiation from the upstream or downstream Met codon41. b | Drosha and Dicer, two RNase III endonuclease family members, are essential for miRNA biogenesis. Drosha and Dicer, as well as cofactors DGCR8 and TRBP, contain one or more dsRBDs. In addition to the catalytic domain RIIID, which is responsible for the RNase III endonucleolytic reaction, unique functional domains, such as the Pro-rich (P) and Arg–Ser-rich (RS) domains, are present in Drosha. By contrast, the DEAD-box RNA helicase, DUF and PAZ domains are present in Dicer. The PAZ domain binds to the 3′ end of miRNAs, whereas the precise role of the DEAD-box RNA helicase domain is unknown. The function of the DUF domain is also unknown. The WW motif of DGCR8 is likely to be involved in protein interactions. Both ADARs and the proteins involved in the miRNA biogenesis pathway bind their dsRNA substrates through dsRBDs. The interaction between dsRNA and dsRBD is not RNA-sequence specific. Therefore, adenosine to inosine (A→I) editing and RNA-interference mechanisms might compete for a common dsRNA substrate, such as primary transcript miRNA (FIGS 6,7). aa, amino acids.