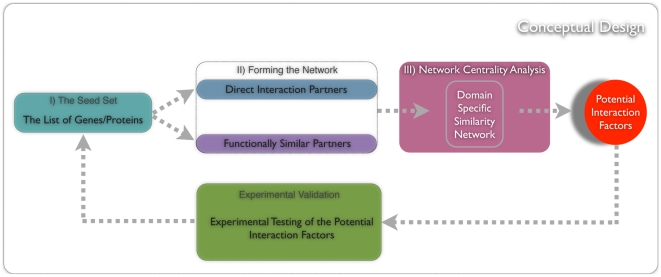
Figure 1. Conceptual design of our prediction framework.
The method consists of three components. I) Compiling “the seed set” from genes/proteins sharing specific characteristic of interest; II) Forming a network by including direct interaction partners described in database(s) and/or functionally similar partners; III) Network centrality analysis on the domain-specific similarity network to obtain potential interaction factors (PIFs). The final step of such an analysis is the experimental validation of PIFs. Confirmed PIFs then can be included in the seed set and the steps I-III can be repeated for identifying new PIFs.