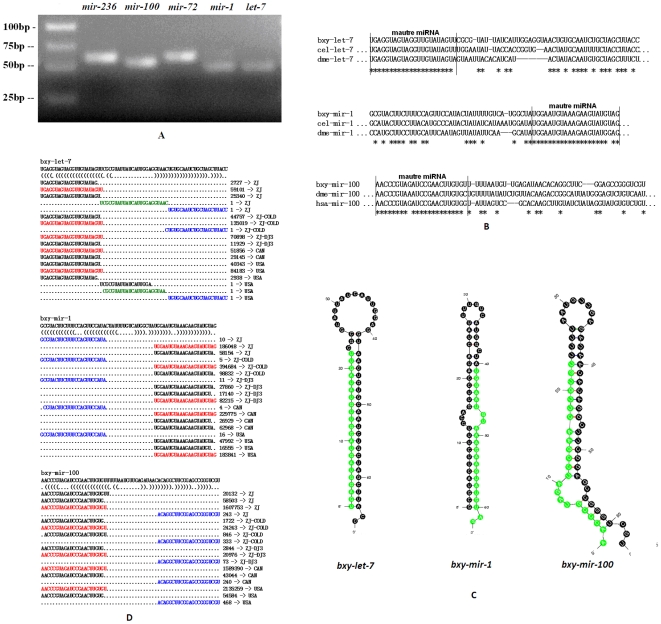
Figure 4. Conserved miRNAs verified in B. xylophilus.
(A) Electrophoretic analysis of conserved miRNA precursor PCR products. (B) Sequence alignment of B. xylophilus miRNAs with known animal miRNAs. (C) Hairpin structures of B. xylophilus let-7, mir-1 and mir-100 miRNA precursors. Nucleotide bases of mature miRNAs are highlighted with green color. (D) Small RNAs derived from the conserved miRNA precursors. The sequence of the let-7, mir-1 and mir-100 hairpins depicted above their bracket-notation secondary structure as determined by RNAfold (Hofacker et al., 2003). Below, each of the small RNA sequences that matched the hairpins is listed, with the number of reads representing each sequence and the corresponding small RNA libraries are shown in the right side. The dominant miRNA sequence is red; the dominant miRNA* species is blue; and the loop-containing sequence is green.