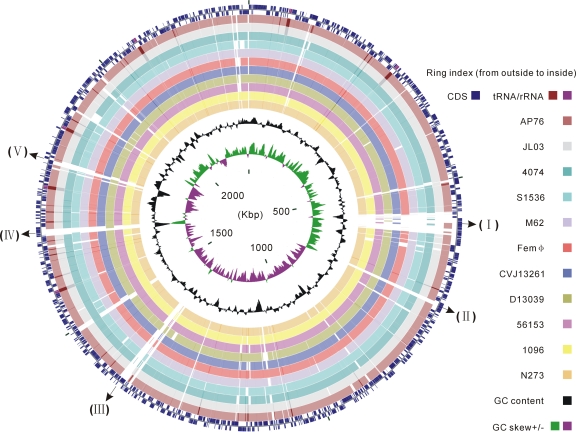
FIG. 1.
Circular representation of sequence conservation between A. pleuropneumoniae serovar 5b strain L20 and 11 strains belonging to different serovars. Circles are numbered from 1 (outermost circle) to 16 (innermost circle). The outermost two circles show CDSs, rRNAs and tRNAs in the L20 genome of A. pleuropneumoniae serovar 5b. The next 11 circles show the coordinates of BLAST hits detected through blastn comparisons (minimum sequence identity of 95% and expected threshold of 10−5) of the L20 reference genome against 11 A. pleuropneumoniae genomes, including two public complete genomes and nine contig sets of new genomes, and each circle is colored according to serovar reference strains: maroon for serovar 7 strain AP76, silver for serovar 3 strain JL03, teal for serovar 1 strain 4074, cyan for serovar 2 strain S1536, light purple for serovar 4 strain M62, red for serovar 6 strain Femϕ, blue for serovar 9 strain CVJ13261, olive for serovar 10 strain D13039, fuchsia for serovar 11 strain 56153, yellow for serovar 12 strain 1096, and orange for serovar 13 strain N273. Overlapping hits appear as darker arcs. The innermost two circles show GC content and GC skew plot of the L20 genome. Several known serovar-specific genomic regions with low sequence identity were numbered as follows: I, the ∼38-kb prophage region; II and III, the coding gene cluster involved in type I restriction-modification system; IV, the LPS O-antigen biosynthesis region; V, the CPS biosynthesis region.