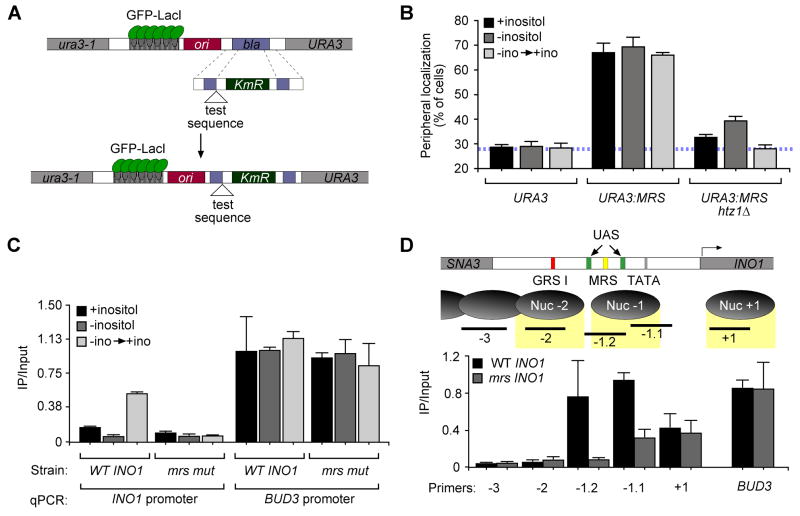
Figure 3. The MRS and the histone variant H2A.Z control peripheral targeting of recently repressed INO1.
A. Scheme for integrating DNA elements for localization experiments (Ahmed et al., 2010). B. Peripheral localization of URA3, URA3:MRS and URA3:MRS in the htz1Δ strain. C. Chromatin immunoprecipitation of HA-H2A.Z (Meneghini et al., 2003) from wild type and mrs mutant INO1strains and quantified using primers to amplify −197 to −284 relative to the INO1 ORF or primers to amplify the BUD3 promoter. D. Top panel: map of nucleosomes in the INO1 promoter. Shown are the positions of GRS I (red box), the MRS (yellow box), the TATA box (grey box), two UASINO elements (green boxes) and the PCR products associated with each nucleosome. Bottom panel: ChIP of HA-H2A.Z from either a wild type or mrs mutant INO1 strain, quantified using primers corresponding to the locations in the top panel or the BUD3 promoter.