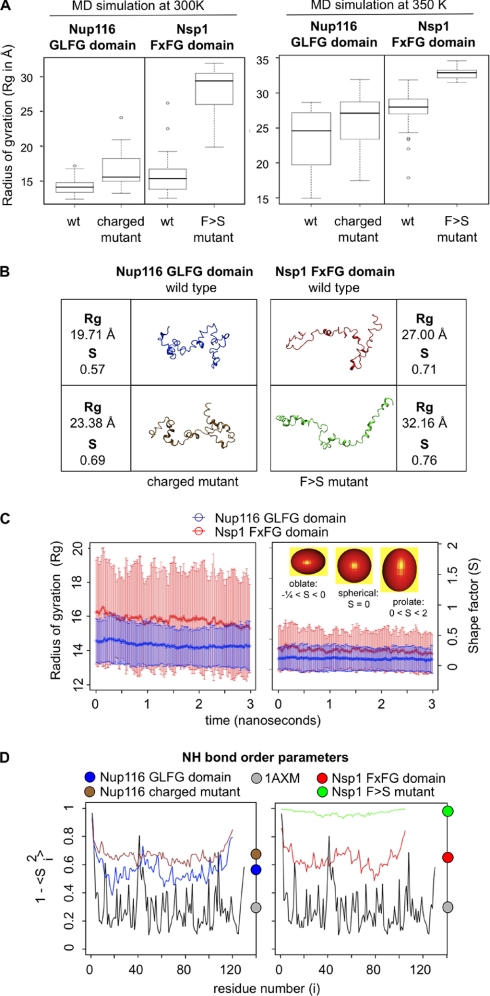
Fig. 4.
Structural dynamics of nup FG domains. A, box plots of the Rg for the FG domain structures simulated. The data represent the mean Rg obtained for 40 independent simulations of each domain at 300 or 350 K. The plots show a box around the central half of the data points enclosing the interquartile range. The error bars (‘whiskers’) enclose the remaining data points except for any outlier points (shown as individual points) that are more than 1.5 times the interquartile range outside the box. B, a representative snapshot of average structures obtained during 350 K simulations. The selected structures were those whose Rg value best matched the measured Stokes radius value and the average S value. The overall best match was with structures in the top of the first quartile in Rg values (i.e. an Rg > 25% of sampled structures; see A). C, plot of the mean and standard deviation (error bars) of the Rg and S values over time for FG domain simulations in implicit solvent at 300 K. Rg provides a measure of compactness for each protein, whereas S provides a statistical measure of its shape. The inset shows ellipsoids with shapes that match sample S parameters. D, order parameter (1 − S2) calculations for FG domains simulated at 300 K. Values were calculated for the amide N–H bond on each residue across all 300 K implicit solvent simulations. High or low values indicate larger or smaller fluctuations, respectively, in the N–H bond angle due to amplitude motions. Values for a folded protein (Protein Data Bank code 1AXM) are shown for comparison. The color circles on the right mark the average value for each protein.