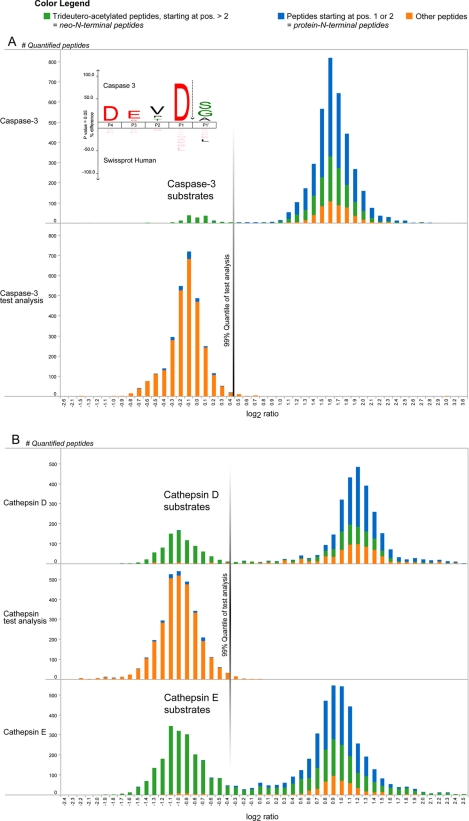
Fig. 2.
Identified human caspase-3 and mouse cathepsin D and E cleavage sites. A, given the experimental design described in the main text, caspase-3-generated neo-N-terminal peptides show light/heavy ratios of about 1 (0 in log2 scale), whereas all other N-terminal peptides hold ratio values distributed around 3 (1.58 in log2 scale) (upper panel). An aliquot of the protease-treated sample analyzed by shotgun proteomics yielded quantified peptides shown in the lower panel, and the border of a one-sided 99% quantile of these peptide ratios was used as ratio cutoff value (this cutoff value was 0.45 and is indicated with a vertical line). The inset shows an iceLogo of all 141 (76 unique) neo-N-terminal peptides generated by caspase-3, which revealed the canonical caspase-3 recognition site DEVD. Further note that many neo-N-terminal peptides with ratios distributed close to 3 are the result of signal peptide removal (as the case for mitochondrial proteins) rather than being generated by contaminating protease activity during caspase-3 incubation. B, freeze-thaw lysates of partially (68% [13C6]Arg) SILAC-labeled primary mouse dendritic cells were incubated with cathepsin D or E. Upon mixing with a control lysate, cathepsin-generated neo-N-terminal peptides have light/heavy ratios of about 0.5 (−1 in log2 scale), whereas all other N-terminal peptides hold ratio values around 2 (1 in log2 scale) (upper and lower panels). A test analysis to determine a ratio cutoff value was performed similarly to the caspase-3 experiment (middle panel; the cutoff value was now −0.31).