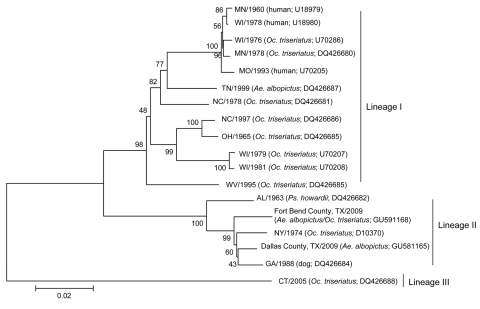
Figure 2.
Phylogeny of La Crosse virus (LACV) medium (M) segment sequences of diverse origins. According to a limited availability of full-length sequences in GenBank, 1,663 nt of the M segment glycoprotein gene open-reading frame are compared. Isolate source and GenBank accession nos. appear after the isolate designation for each taxon. Sequences were aligned by ClustalW (10) and neighbor-joining and maximum-parsimony trees were generated by using 2,000 bootstrap replicates with MEGA version 4 software (10). Highly similar topologies and confidence values were derived by all methods and a neighbor-joining tree is shown. Scale bar represents the number of nucleotide substitutions per site. The 2009 Texas (TX) isolates group with strong support with lineage 2 viruses of the extreme south and New York (NY), which suggests a likely southern origin for LACV isolates. MN, Minnesota; WI, Wisconsin; Oc., Ochlerotatus; MO, Missouri; TN, Tennessee; Ae., Aedes; NC, North Carolina; OH, Ohio; WV, West Virginia; AL, Alabama; Ps., psorophora; GA, Georgia; CT, Connecticut.