(
a) is a cartoon depicting the
Drosophila circadian molecular network; protein and mRNA are represented by capital letters and lower case, respectively. Red arrows and cyan blocked lines indicate stimulatory and inhibitory interactions, respectively. The green arrow ending in X indicates that CRY protein enhances the degradation of TIM. (
b) and (
c) plot recurring orbits of the wt and
cwo-mutant models in LD (cycles 100 to 120000), respectively; observe the jitter/variation when CWO is absent. The variability of CWO is proportional to the variability of each direct target gene at the times of its peak and trough (
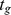
, see
File S1):
(
d) plots the variability of CWO (y-axis) vs.
per mRNA (
x-axis) at the peak-time of
per. (
e) illustrates the theoretical results predicting that the jitter of CWO dampens the jitter of direct targets at the times of their peaks and troughs. (
f)–(
k) plot
per oscillations in simulations where the CLK/CYC of each cycle (total = 70) of the wt (
f–h) and the
cwo-mutant (
i–k) models is pulsed at ZT = 14 hr by pseudorandom numbers drawn from a uniform distribution on [0, 0.001] (
f and
i), [0, 0.01] (
g and
j), and the unit interval (
h and
k), respectively. The unit of the y-axes of
b–c and
f–k is arbitrary.
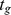 , see File S1):
, see File S1):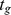 , see File S1):
, see File S1):