Table 1. Summary of crystal parameters, data-collection and refinement statistics for YP_001813558.1 (PDB code 3nl9).
Values in parentheses are for the highest resolution shell.
| λ1 MADSe | λ2 MADSe | |
|---|---|---|
| Crystal parameters | ||
| Space group | C2 | |
| Unit-cell parameters (Å, °) | a = 52.09, b = 69.04, c = 50.21, β = 111.8 | |
| Mosaicity (°) | 0.91 | |
| Data collection | ||
| Wavelength (Å) | 1.0000 | 0.9798 |
| Resolution range (Å) | 39.6–1.78 (1.83–1.78) | 39.6–1.78 (1.83–1.78) |
| No. of observations | 43073 | 43110 |
| No. of unique reflections | 15531 | 15528 |
| Completeness (%) | 98.1 (97.9) | 98.1 (97.3) |
| Mean I/σ(I) | 9.8 (2.1) | 8.6 (1.8) |
| Rmerge on I† | 0.069 (0.555) | 0.082 (0.563) |
| Rmeas on I‡ | 0.086 (0.687) | 0.102 (0.698) |
| Rp.i.m. on I§ | 0.050 (0.401) | 0.059 (0.408) |
| Overall B factor from Wilson plot (Å2) | 21.3 | 21.0 |
| Model and refinement statistics | ||
| Data set used in refinement | λ1 MADSe | |
| Resolution range (Å) | 39.6–1.78 | |
| No. of reflections (total) | 15531 | |
| No. of reflections (test) | 788 | |
| Completeness (%) | 97.8 | |
| Cutoff criterion | |F| > 0 | |
| Rcryst¶ | 0.177 | |
| Rfree†† | 0.222 | |
| Stereochemical parameters | ||
| Restraints (r.m.s.d. observed) | ||
| Bond angles (°) | 1.30 | |
| Bond lengths (Å) | 0.015 | |
| Average protein isotropic B value (Å2) | 26.0‡‡ | |
| Average solvent isotropic B value (Å2) | 33.6 | |
| ESU§§ based on Rfree (Å) | 0.14 | |
| Protein residues/atoms | 169/1340 | |
| Water/cryoprotectant molecules | 141/2 | |
R
merge = 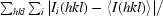
 .
.
The redundancy-independent (multiplicity-weighted) merging R factor, R
meas = 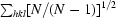
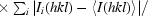
 (Diederichs & Karplus, 1997 ▶).
(Diederichs & Karplus, 1997 ▶).
The precision-indicating merging R factor, R
p.i.m. = 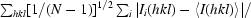
 (Weiss & Hilgenfeld, 1997 ▶; Weiss et al., 1998 ▶).
(Weiss & Hilgenfeld, 1997 ▶; Weiss et al., 1998 ▶).
R
cryst = 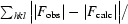
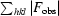 , where F
calc and F
obs are the calculated and observed structure-factor amplitudes, respectively,
, where F
calc and F
obs are the calculated and observed structure-factor amplitudes, respectively,
R free is the same as R cryst but for 5.1% of the total reflections chosen at random and omitted from refinement
This value represents the total B and includes both TLS and residual B components.
